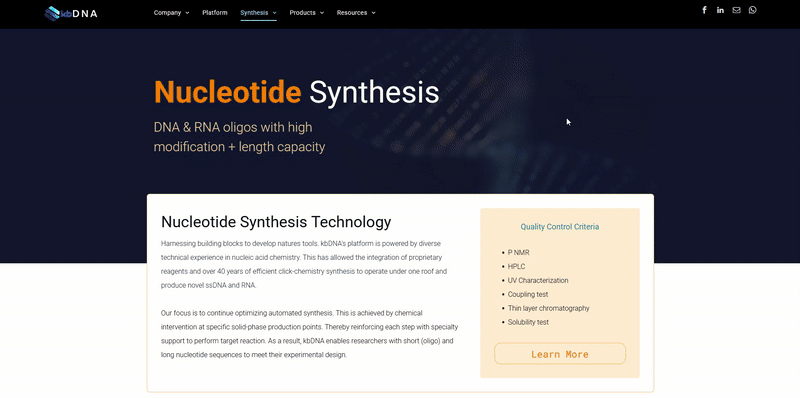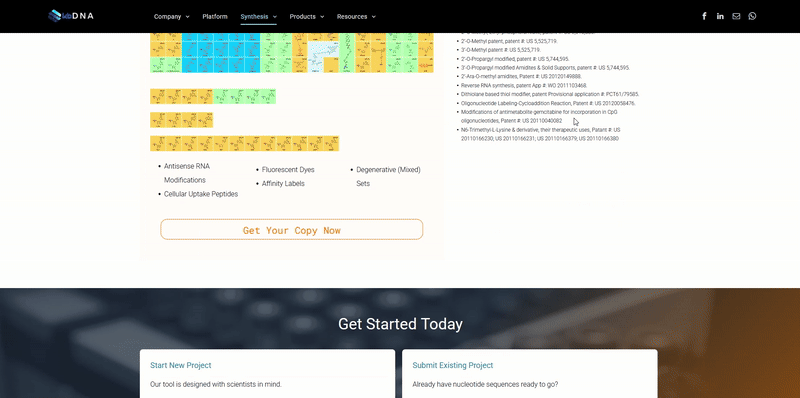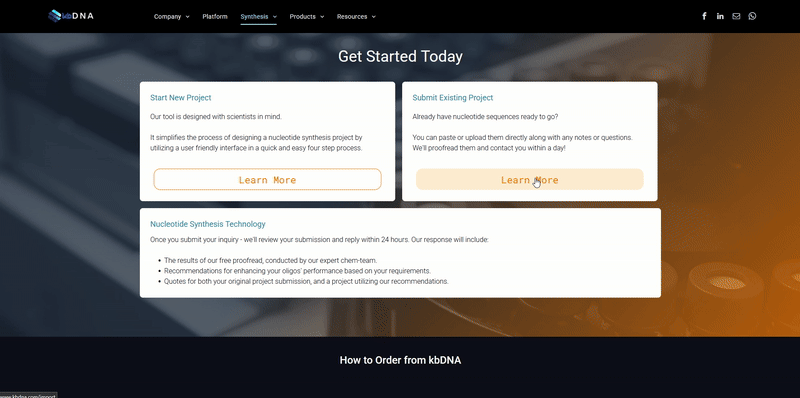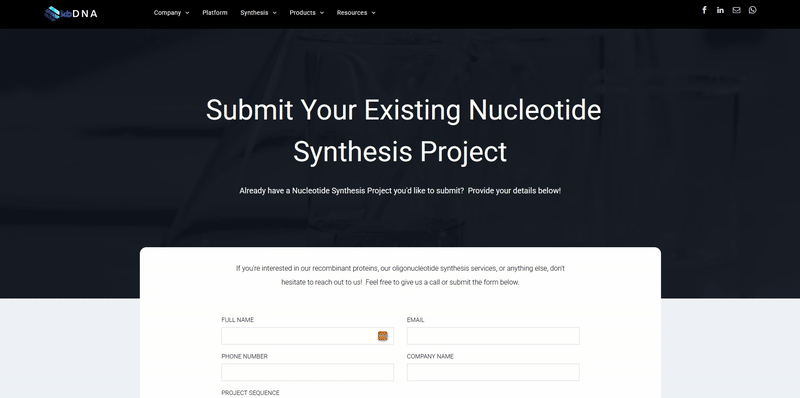
Obsessing Over CRISPR Isn’t Making It Any Safer, but RNA Might
Obsessing Over CRISPR Isn’t Making It Any Safer, but RNA Might
Overcoming the challenges of off-targeting with synthetic guide RNA
Every now and then, a breakthrough discovery can be significant enough to introduce new technology that disrupts traditional scientific research. These discoveries are often easy to identify by their ability to dominate headlines and attract obsessive attention from all audiences. With respect to this article, I am referencing the groundbreaking discovery of Clustered Regularly Interspaced Short Palindromic Repeats or CRISPR for short. Not to say that the attention generated by CRISPR is not entirely deserved. If anything, CRISPR/Cas9 and its technology has proven a very innovative molecular tool toward various lab applications. CRISPR is coined the ‘cute & paste’ of gene-editing (as if it were that easy) and though the technology has been very progressive, it’s limitations continue to hang over our heads. To put it simply, the CRISPR-associated protocols are riddled with errors. They have to be run repeatedly and tweaked to reduce the principal obstacle; off-targeting. (thunder clap!)
Off-target genome editing refers to nonspecific and unintended genetic modifications that can arise through the use of engineered nuclease technologies such as CRISPR/CAS9. Previously, we've seen off-targeting effects cause a major set-back to a generation of CAR-T potential. However, in the case of off-targeting with prior models of CAR-T; the issue was random binding to healthy surface protein (antigens) - similar to the ones being targeted on hard tumors. With CRISPR, the off-targeting effect is a little more complex. To better understand it, let us revisit the fact that the CRISPR/CAS9 system is directed to its target DNA region via an attached nucleic acid sequence, referred to as the “guide RNA (gRNA)”. This gRNA is designed to bind to a specific area along the DNA, complementary to its sequence. Once successfully (or unsuccessfully) attached, the CRISPR/Cas9 gene-editing mechanism initiates a base; deletion, mutation, or introduction. If this is done in the incorrect location, side effects are limitless. This has been a recurring issue in both class I and II CRISPR systems. Several off-targeting solutions are being trialed, which include the use of multiple protein supports and digging into the other proteins of the Cas family. However, no solution shows greater promise than optimizing the single guide RNA (sgRNA) design.
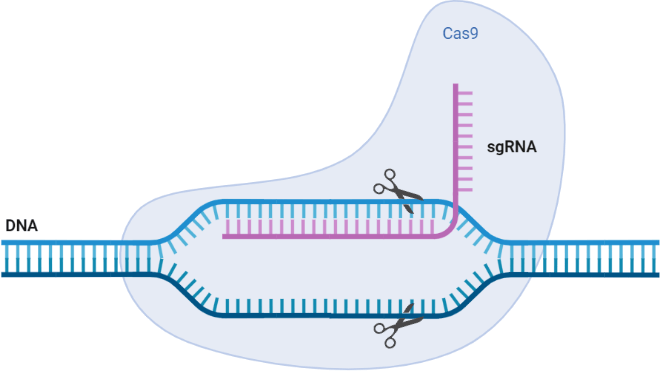
Fig 1. This base template provides a general overview of a CRISPR/Cas9 system for genome editing. Cas9 protein cleaves a specific DNA sequence guided by sgRNA.
This approach does not require adding more components to the process and is centered around specificity - thereby limiting incompatibility which is a recurring issue with other CRISPR off-targeting solutions. The gRNA design is not exactly a new approach. However, the employment of more rational design and extending engineering to its secondary structure; introduces new capabilities of applying hairpin formations, which can be attached to the 5’ end of the sgRNA (hp-sgRNA). The resulting hairpin structure could then function as a support barrier to an R-loop formation. Notably, this offers a kinetic model of R-loop design, that can provide more specificity to the on-target site and regulate more against the off-target ones.
...employment of more rational design and extending engineering to its secondary structure; introduces new capabilities of applying hairpin formations
A recent study at Duke University tested their own version of this methodology with significant results. Following their successful synthesis of the concept RNA structure and functional experimentation; the research team assessed the effects on multiple Cas variants including; spCas9 and saCas9. The study did leave a few questions unanswered, such as the consistency of this approach or the influence of varying conditions relative to R-loop stability. Not to mention, the ability to synthesize RNA structures to meet such novel design is strenuous in itself - and one that has been challenging different areas of therapeutic RNA (RNAi) for some time now. Regardless, the data being collected with this approach is definitely progress and the more we focus on building off of it and less on sci-fi applications of CRISPR, the better we are. Unfortunately, science fiction seldom converts into therapeutic application.
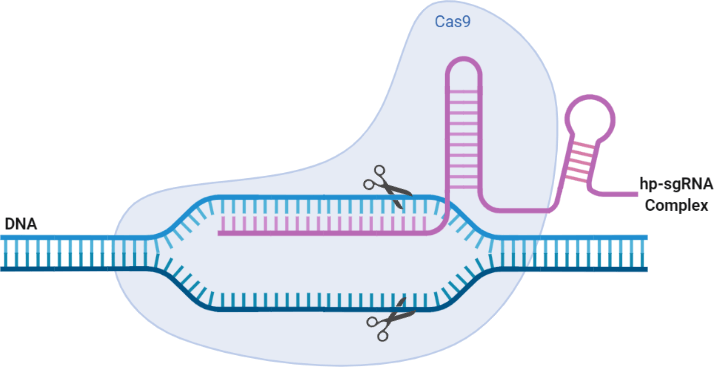
Fig 2. This schema represents an overview of the CRISPR/Cas9 system with the incorporation of a hairpin formation in the secondary structure of the sgRNA
To do our part; we try and relieve some of the manufacturing challenges by further considering stability and structural components, when reviewing RNA sequence submissions, prior to synthesis. As part of our standard proofreading service, we offer this consultation at no-cost and we encourage other manufacturers to incorporate a similar approach to help keep the traction going for the use of RNA against off-targeting.
References
- Kocak, D. D., Josephs, E. A., Bhandarkar, V., Adkar, S. S., Kwon, J. B., & Gersbach, C. A. (2019). Increasing the specificity of CRISPR systems with engineered RNA secondary structures. Nature Biotechnology.
- Wright, A. V., Nunez, J. K. & Doudna, J. A. Biology and applications of
CRISPR systems: harnessing Nature’s toolbox for genome engineering. Cell 164, 29–44 (2016). - Shmakov, S. et al. Discovery and functional characterization of diverse class 2 CRISPR–Cas systems. Mol. Cell 60, 385–397 (2015).
- Burstein, D. et al. New CRISPR–Cas systems from uncultivated microbes. Nature 542, 237–241 (2017).
- Barrangou, R. & Doudna, J. A. Applications of CRISPR technologies in research and beyond. Nat. Biotechnol. 34, 933–941 (2016).
- Fig1. & Fig2 Created with BioRender.com