Can the Application of siRNA Help Improve Our Targeting of Coronavirus?

Can the Application of siRNA Help Improve Our Targeting of Coronavirus?
Here’s what research has learned so far
As the race for the COVID19 vaccine continues to dominate headlines, we wanted to try and get ahead with a topic that will inevitably become a major focus for those developing antiviral therapies. That topic is centered around the challenges in targeting a coronavirus’ polyprotein structure with high selectivity. This is due to several factors such as the virus’ genomic size, its sequence mutation rate and in-vitro experimental limitations. Consequently, increasing the demand for strategic approaches and creative tools for a laboratories design of experiments. In this review, we make a case for utilizing short-interfering RNA (siRNA) tools and antisense or RNA interference (RNAi) technology as potential solutions to the coronavirus targeting challenges along with highlighting key considerations for identifying viral structural regions.
The Virus
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) or COVID-19 has been referenced by many via its various synonyms and some inaccurate names. This has caused a bit of confusion with audiences who aren’t too familiar with viral taxonomy. For the sake of consistency, we will stick with COVID19. COVID19 belongs to the coronaviridae family in the nidovirales order and is one of the largest RNA viruses with a genomic makeup of almost 30 kb1. Although COVID19 is characteristically distinct from other coronaviruses, its viral encoded genes and their assembly conform to the shared principles. The primary gene order from 5’ to 3’ follows; ORF1a + ORF1b (ORF1ab), spike (S), envelope (E), membrane (M) and nucleocapsid (N) proteins. ORF1ab encodes most of the viral regulatory proteins for transcription and translation2. Next the virus’ primary structural proteins; S, M, E form the lipid bilayer while N proteins build the helical nucleocapsid3. In addition, approximately eight accessory or group specific genes are interspersed between these structural proteins2,4. A visual representation of COVID19’s genomic organization can be found in the following schema (Fig. 1).
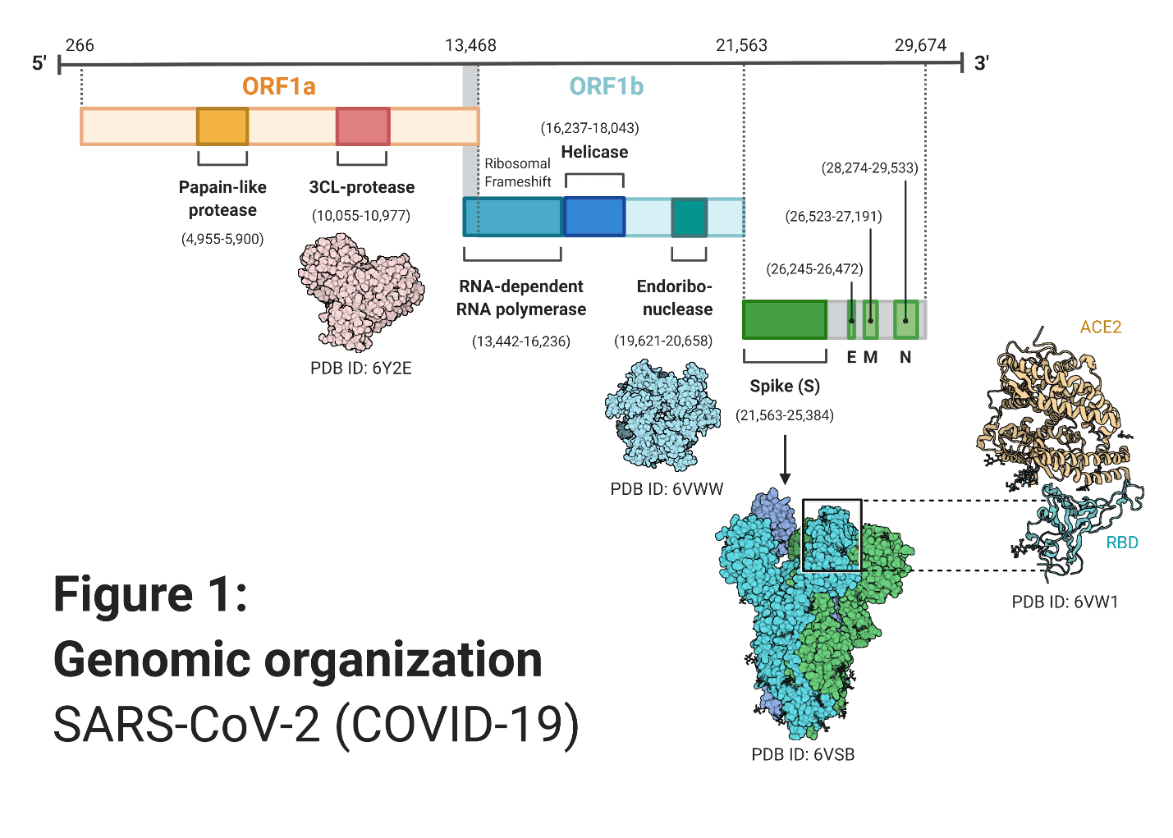
Fig 1:
Genomic Organization SARS-CoV-2 (COVID-19)
Targeting Structural Regions
Proteomic analysis suggests that the interactions between structural proteins present attractive opportunities for anti-SARS agent development5,6. Therefore, the next step would be individually analyzing each domain and determining optimal regions for targeting. Since RNA viruses are variable due to their replications high mutation rate, the underlying challenge in analyzing their structural genes - involves identifying consistent regions with low or complying sequence mutation rates. Published data on coronavirus’ E and N genes suggests that siRNA targeting their proteins leader sequence, terminal repeat short (TRS) and 3’-untranslated (3’-UTR) regions could effectively inhibit the expression of these targets14. However, current literature focus and reproducibility has not been strong enough to support them as top candidates.
Instead- in this review, we evaluate the two structural proteins, S and M in the context of siRNA antisense qualifying standards.
(S) Spike Proteins
The spike is a glycoprotein surface projection which is critical for viral attachment and entry into host cells. Variations of S protein throughout strains of coronavirus have also shown it to be responsible for host range and tissue tropism7. Results from prior genetic sequencing of patients with similar SARS virus have shown significant variation of S gene in the viral strain. The S1 subunit of S protein is the major antigenic moiety for coronaviruses33. Therefore, as the virus evolves in host environments; it is shown to be prone to having high mutation rates33. Notably, cases have been made for S and S1 domain as ACE2 blocking regions for monoclonal antibody development. It is also worth noting that articles have reported the possibility of inhibiting replication by targeting S gene at U6 promoter site using short-hairpin RNA (shRNA)16. Although this finding does support the application of RNAi, the technical and safety differences between shRNA and siRNA does not directly transfer and demands further exploration. Thus, in principle; S protein would be a potential domain for investigating targeting regions for siRNA compounds.
(M) Membrane Protein
The membrane protein is a triple-spanned transmembrane protein. M gene codes for the most abundant of all the structural proteins and plays an essential role for viral assembly10. The specifics of assembly such as sequence of co-expressing interactions and minimal criteria for viral like particle (VLP) assembly are still controversial for COVID19. As a reference, we can use similar studies of SARS-CoV which indicate that either combination of M & N or M & E are enough to form and release VLPs. Contrary, others showed that N and E, both must co-express with M; in order to efficiently produce and release VLP5,6. While we continue to refine our understanding of the full series of interactions, the current knowledge is enough to highlight the comprehensive role of M protein while influencing the focus on it during analysis.
An attractive feature of M protein is its interactions with intracellular signaling pathways. SARS M protein has been shown to inhibit the nuclear factor kappa B (NF-kB) pathway through direct contact with upstream kinase IKKB11. It can also inhibit dsRNA-induced interferon B production by disrupting the formation of the TRAF3-TANK-TBK1/IKKE complex12. Signaling blockades such as these can significantly impair both the hosts innate and adaptive immune responses. Many researchers would agree that this is a clear opportunity for an inhibition strategy. Ultimately, supporting M gene as a high potential targeting candidate for anti-SARS drug development.
Application of siRNA
Site-specific inhibition of cellular mRNA by antisense technology can be achieved in mammalian cells by using 21-25 nucleotide (nt) duplexes of synthetic RNA. This category of synthetic RNA is referred to as siRNA and the discovery of their RNAi utility has been successfully adopted by the infectious disease community for a variety of viruses. This has been the case for inhibiting expression and replication in; poliovirus, HIV-1, HCV, Hepatitis B and influenza virus. Therefore, it only makes sense that siRNA will be explored for RNAi strategies against COVID19.
The complexity of antisense technology, particularly RNAi can be simplified as a two-step process. First, long double-stranded RNA (dsRNA) is processed by the RNAse III enzyme, Dicer into the intended 21-25 nt siRNA. Alternatively, siRNA can be introduced directly via RNA-induced silencing complex (RISC). Secondly, the RISC system ultimately pairs the siRNA by means of antisense to its target messenger RNA (mRNA) complimentary sequence. RISC then proceeds to degrade the target mRNA at the non-paired siRNA sites. To understand how this translates into the research setting, its helpful to analyze previously published work on similar SARS viruses and RNAi applications.
For example, studies developing siRNA to the S gene have reported that targeting the genomeat S protein and NSP12-coding regions has shown success in silencing SARS-CoV expression in cell culture, in-vitro13. Further evaluation of efficacy in rhesus macque models, found that this approach decreased viral presence, provided relief from infection-induced fever and reduced acute diffuse alveolar damage13.
Other findings strongly support siRNA targeting of the M gene. It has been demonstrated that cross-analyzing M gene sequences from 15 different SARS-CoV isolates, allowed the identification of two novel conserved targeting regions for siRNA testing14. Real-time quantitative RT-PCR (qRT-PCR) testing of the regions confirmed that siRNA targeting the 3’ half of M gene (si-M2) and the 5’ half (si-M1) both significantly downregulated M gene mediated upregulation of interferon B expression. With si-M2 having induced higher inhibition potency14. Similar findings have also helped support the theory that M gene-specific siRNA might function in a sequence-dependent manner.
The Technical Reality
The innovations in RNAi and its application of siRNA targeting viruses shows clear potential. However, it is imperative to understand that the laboratory research behind this is very rigorous and technically demanding. Those familiar with the methods involved will be the first to emphasis the high degree of experimental analysis required throughout all the steps in developing and testing siRNA. Not to say that this isn’t the case with other research applications, but there is a significantly higher level of consideration for efficacy and safety when planning to transfer this technology into a clinical setting. Not to mention the conflicting relationships between key data such as chemical concentration and toxicology. However, that topic is a rabbit hole of its own. To keep it relevant and help provide an understanding of the technical methodology; we outlined the common experiment workload for the research analysis of siRNA (Fig. 2).
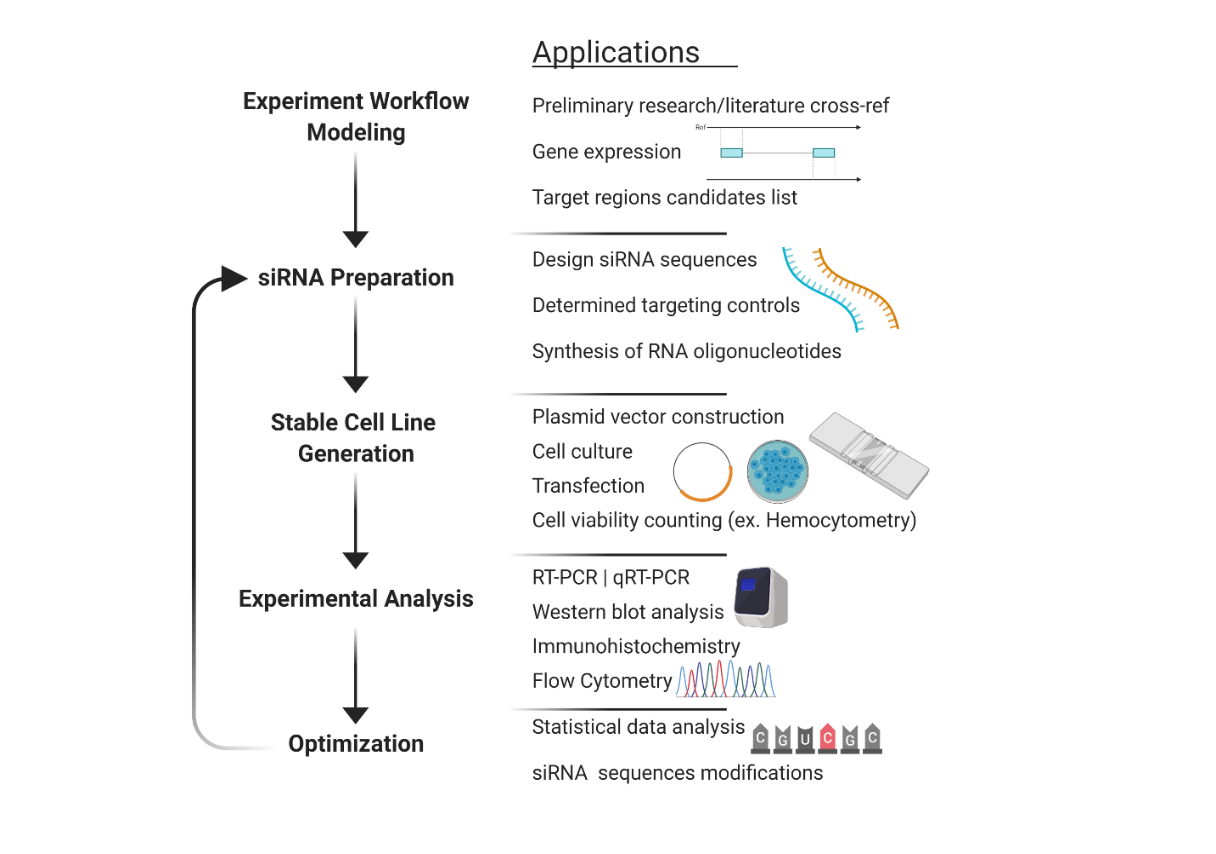
Fig 2:
Schematic representation of the general research process in early development of siRNA compounds-specific to target molecule regions. The left-side process displays the sequence of objectives and listed on the right, are the applications relevant to each stage.
What makes this research process even more challenging is its application toward a novel or complex virus. As mentioned, viruses like SARS mutate, and their structures operate in sophisticated fashion. Therefore, thorough analysis is required to go beyond observation to also help identify opportunities for introducing methodologies. In the case of antisense targeting COVID19, each structural domain must undergo individual evaluations to qualify for candidate regions. Following, siRNA sequences are designed against those regions of interest. Then both sequences and regions are tested for validations and optimization is pursued accordingly. Throughout this principled approach, discovery of complicating factors is anticipated along with demand for creative intervention. This has been the case with similar coronaviruses, particularly SARS-CoV which resulted in an array of findings to consider in experiment protocols.
These findings include:
- Human coronaviruses are difficult to culture in-vitro. However, their direct cytopathic effects can be demonstrated by inoculating viral isolates into Vero E6 cells – making the cells model suitable enough for studying the effects of siRNA15.
- Beyond the study of siRNA properties and aiding sequence alignment, Vero E6 cells are limited in downstream value. Anything more comprehensive requires cell lines derived from preferably mammalian. It is suggested that human embryonic kidney cell line (HEK293) and SV40 T transformed HEK293 (HEK293T) are the most stable for this SARS-CoV application14.
- The incorporation of fluorescent control protocols into flow cytometry analysis is strongly recommended for thorough expression inhibition analysis and tracking benchmark15.
- The use of partial regions of viral proteins in assay validation are shown to be unreliable and non-compliant with commercial antibodies. Native donor purified protein is the standard. However, they aren’t always an option. The closest alternative would ideally be full sequence whole recombinant protein, untagged9.
- Thermodynamic properties will play a large role in modifying siRNA design, particularly in the sense strand. In one case, functional siRNA duplexes displayed a lower internal stability at 5’ antisense terminal base pair end than the non-duplexes. This prompted changes in nucleotides at certain positions of the sense strand (that expressed low inhibition activity); to unpair the 5’ end of the antisense strand. This allowed the siRNA to reduce target gene expression more effectively15.
Discussion
At present, several potential SARS therapies are under development such as vaccines, treatments and interferons. In terms of COVID19, the urgent priority in the market respectively are vaccines. However, what we have learned from pandemics is that coronaviruses are packed with uncertainty. Whether they are within their structural chemistry, proteomics or behavioral features (ex. symptoms) - this class of virus requires a combination of therapies to help strategically tackle it medically. This demands research to innovate our toolkit with sharper solutions for better precision medicine. One essential solution that correlates with medical efficacy, is the level of selectivity our drugs can target these viruses. Due to being highly customizable and versatile; the application of antisense RNAi using modified siRNA present a made-to-measure opportunity for improved targeting of coronavirus structures.
As explained, coronaviruses present various research challenges such as sequence variability during in-vitro analysis. Of similar importance is the difficulty producing proof of efficacy and safety in the in-vivo setting. Despite their published potential and manufacturing capability, it is important to understand that siRNA compounds come with their own drawbacks as well. Making synthetic RNA clinically safe is very labor intensive and requires a very elaborate biochemistry strategy. siRNA compounds are also transient and thereby limited as core mechanisms in an infectious disease drug. Thus, one would theorize that its targeting advantages are more of a companion technology than a comprehensive solution. As is everything in scientific research, further exploration and validation is required on several components of the topic. In order to do so, it is important to consider a lot of what has already been observed and simultaneously follow similar research currently in-progress. The contents of this article are intended to support guiding readers with the former.
References
- Masters, P.S. The molecular biology of coronaviruses. Advan. Virus Res. 2006, 66, 193-292.
- Frieman, M.; Baric, R. Mechanisms of severe acute respiratory syndrome pathogenesis and innate immunomodulation. Microbiol. Mol. Biol. Rev. 2008, 72, 672-685.
- Perlman, S.; Dandekar, A.A. Immunopathogenesis of coronavirus infections: implications for SARS. Nat. Rev. Immunol. 2005, 5, 917-927.
- Narayanan, K.; Huang, C.; Makino, S. SARS coronavirus accessory proteins. Virus Res. 2008, 133, 113-121.
- Huang, Y.; Yang, Z.Y.; Kong, W.P.; Nabel, G.J. Generation of synthetic severe acute respiratory syndrome coronavirus pseudoparticles: implications for assembly and vaccine production. J. Virol. 2004, 78, 12557-12565.
- Hsieh, P.K.; Chang, S.C.; Huang, C.C.; Lee, T.T.; Hsiao, C.W.; Kou, Y.H.; Chen, I.Y.; Chang, C.K.; Huang, T.H.; Chang, M.F. Assembly of severe acute respiratory syndrome coronavirus RNA packaging signal into virus-like particles is nucleocapsid dependent. J. Virol. 2005, 79, 13848-13855.
- Kuo L, Godeke GJ, Raamsman MJ, Masters PS, Rottier PJ.Retargeting of coronavirus by substitution of the spike glycoprotein ectodomain: crossing the host cell species barrier. J Virol 2000; 74:1393-406.
- Ruan YJ, Wei CL, Liu ET, et al. Comparative full-length genome sequence analysis of 14 SARS coronavirus isolates and common mutations associated with putative origins of infection. Lancet 2003; 361:1779-85.Y39 9
- Ruan YJ, Wei CL, Liu ET, et al. Comparative full-length genome sequence analysis of 14 SARS coronavirus isolates and common mutations associated with putative origins of infection. Lancet 2003; 361:1779-85.
- Stertz, S.; Reichelt, M.; Spiegel, M.; Kuri, T.; Martinez-Sobrido, L.; Garcia-Sastre, A.; Weber, F.; Kochs, G. The intracellular sites of early replication and budding of SARS-coronavirus. Virology 2007, 361, 304-315.
- Fang, X.; Gao, J.; Zheng, H.; Li, B.; Kong, L.; Zhang, Y.; Wang, W.; Zeng, Y.; Ye, L. The membrane protein of SARS-CoV suppresses NF-kappaB activation. J. Med. Virol. 2007, 79, 1431-1439.
- Siu, K.L.; Kok, K.H.; Ng, M.H.; Poon, V.K.; Yuen, K.Y.; Zheng, B.J.; Jin, D.Y. Severe acute respiratory syndrome coronavirus M protein inhibits type I interferon production by impeding the formation of TRAF3.TANK.TBK1/IKKepsilon complex. J. Biol. Chem. 2009, 284, 16202-16209.
- Li B-j. Tang Q. Cheng D et al. Using siRNA in prophylactic and therapeutic regimens against SARS coronavirus in rhesus macaque. Nature Med. 11 2005, 944-951
- Wang, Y.; Cao, Y.-L.; Yang, F.; Zhang, Y.; Wang, S.-H.; Liu, L. Small Interfering RNA Effectively Inhibits the Expression of SARS Coronavirus Membrane Gene at Two Novel Targeting Sites. Molecules 2010, 15, 7197-7207.
- SHI, Y., YANG, D., XIONG, J. et al. Inhibition of genes expression of SARS coronavirus by synthetic small interfering RNAs. Cell Res 2005, 15, 193–200.
- Zhang Y, Li T, Chang Z, et al. Silencing SARS-CoV Spike protein expression in cultured cells by RNA interference. FEBS Lett 2004; 560:141-6.