An Overview of Linkers for Recombinant Fusion Proteins
An Overview of Linkers for Recombinant Fusion Proteins
The selection or rational design of a linker to join fusion protein domains is an important, under-explored area in recombinant fusion protein technology
Introduction
A fusion protein is a protein consisting of at least two domains that are encoded by separate genes that have been joined so that they are transcribed and translated as a single unit, producing a single polypeptide. By genetically fusing two or more protein domains together, the fusion protein product may obtain many distinct functions derived from each of its component moieties. Three of the most frequent uses of fusion proteins are: as aids in the purification of cloned genes, as reporters of gene expression levels, and as histochemical tags to enable visualization of the location of proteins in a cell, tissue, or organism. More recent applications of the fusion protein technology also include creating novel protein therapeutics and improving the performance of current protein drugs.
Two indispensable elements are required for the successful construction of a recombinant fusion protein: the component proteins and linkers. In most cases, the choice of the component proteins is relatively straightforward as it is based on the desired functions of the fusion protein product. However, the selection of a suitable linker(s) to join the protein domains together can be complicated and is often neglected in the design of fusion proteins. Direct fusion of functional domains in the absence of a linker may lead to many undesirable outcomes, including misfolding of the fusion proteins, low yield in protein production, or impaired bioactivity. Therefore, the selection or rational design of a linker to join fusion protein domains is an important, yet under-explored, area in recombinant fusion protein technology.
Summarized below is the current knowledge of linker properties, design and functionality in recombinant fusion protein technology.
Linkers from Naturally Occurring Multi-Domain Proteins
Similar to recombinant fusion proteins, naturally-occurring multi-domain proteins are composed of two or more functional domains joined by linker peptides. These linker peptides serve to connect the protein moieties, and also carry out many other functions, such as maintaining cooperative inter-domain interactions and preserving biological activity. Two studies by Argos (1) and George and Heringa (2) have independently compared several properties of natural linkers, such as length, hydrophobicity, amino acid residues, and secondary structure. The results of these studies are summarized in Table 1.
Table 1: Properties of linkers derived from natural proteins
| Property | Argos (1) | George and Heringa (2) | |
|---|---|---|---|
| Length (Number of amino acid residents) | 6.5 | 10.0 ± 5.8 (small: 4.5 ±.7; medium: 9.1 ± 2.4; large: 21± 7.6) | |
| Hydrophobicity (3) | -- | 0.65 ± 0.09 (small: 0.69 ± 0.11; large: 0.62 ± 0.08) | |
| Amino acid propensity (4) | Thr | 1.55 | 1.017 |
| Amino acid propensity (4) | Ser | 1.46 | 0.947 |
| Amino acid propensity (4) | Pro | 4.35 | 1.299 |
| Amino acid propensity (4) | Gly | 1.25 | 0.835 |
| Amino acid propensity (4) | Asp | 1.25 | 0.916 |
| Amino acid propensity (4) | Lys | 1.16 | 0.944 |
| Amino acid propensity (4) | Gln | 1.13 | 1.047 |
| Amino acid propensity (4) | Asn | 1.09 | 0.944 |
| Amino acid propensity (4) | Ala | 1.05 | 0.964 |
| Amino acid propensity (4) | Val | 1 | 0.955 |
| Amino acid propensity (4) | Glu | 0.87 | 1.051 |
| Amino acid propensity (4) | Arg | 0.84 | 1.143 |
| Amino acid propensity (4) | Ile | 0.81 | 0.922 |
| Amino acid propensity (4) | Tyr | 0.75 | 1 |
| Amino acid propensity (4) | Met | 0.75 | 1.032 |
| Amino acid propensity (4) | Phe | 0.69 | 1.119 |
| Amino acid propensity (4) | His | 0.55 | 1.014 |
| Amino acid propensity (4) | Cys | 0.35 | 0.778 |
| Amino acid propensity (4) | Trp | 0.23 | 0.895 |
| Amino acid propensity (4) | Leu | NA | 1.085 |
| Secondary Structure (5) | α-Helical | 13% | 38.3% (small: 21%; large: 31.4%) |
| Secondary Structure (5) | β-Strand | 12% | 13.6% (small: 33.6%; large: 10.4%) |
| Secondary Structure (5) | Coil | 59% | 37.6% (small: 36.9%; large: 45.4%) |
| Secondary Structure (5) | Turns | 16% | 8.4% (small: 8.5%; large: 12.8%) |
| # | Description |
|---|---|
| 1 | Data taken from study by Argos [23] |
| 2 | Data taken from study by George and heringa [24] |
| 3 | Hydrophobicity values taken from Eisenberg's normalized consensus residue hydrophobicity scale, which ranges from 0 (hydrophilic) to 1 (hydrophobic) |
| 4 | Calculated from the ratio of a single amino acid occurrence in the linker et compared to its occurrence in the full protein set, where values greater than 1 (shaded) indicate larger than average occurrences in linker sequences) |
The main findings of these two studies are:
- Structural environment of linkers: These studies calculated the average normalized solvent accessibility and hydrophobicity of various linkers. Higher solvent accessibility was observed with increasing length of linkers, suggesting that longer linkers were more likely to be exposed to the solvent. Consistent with this finding, the average hydrophobicity of the linkers decreased with increase in their length, indicating that longer linkers were more hydrophilic and therefore more exposed to the aqueous solvent than shorter linkers.
- Amino acid residue preferences: These studies also determined the amino acid preference in natural linkers by calculating the ratio of the occurrence of a single amino acid in the linker vs the full protein (Table 1). Values greater than 1 indicate a higher occurrence of a particular amino acid in the linker. Threonine (Thr), serine (Ser), proline (Pro), glycine (Gly), aspartic acid (Asp), lysine (Lys), glutamine (Gln), asparagine (Asn), and alanine (Ala) were identified as preferable linker constituents by Argos (1), whereas Pro, arginine (Arg), phenylalanine (Phe), Thr, glutamic acid (Glu) and Gln were preferred in the George and Heringa study (2). In general, preferable amino acids were polar uncharged or charged residues, which constitute approximately 50% of naturally encoded amino acids. Both studies suggested that Pro, Thr, and Gln were the preferable amino acids for natural linkers. Among them, Pro is a unique amino acid with a cyclic side chain which causes a very restricted conformation.
- Secondary structures of linkers: Natural linkers adopt various secondary structures, such as helical, β-strand, coil/bend and turns, to exert their functions. From George and Heringa’s analysis, most linkers on average exhibited α-helix (38.3%) or coil/bend (37.6%) secondary structures (Table 1). The conformations were slightly changed when small vs. large linkers were compared, where the majority of linkers adopted coils. On the other hand, the study by Argos showed that the majority of the linkers adopted coil structures (59%).
Overall, natural linkers primarily adopted extended conformations, and had independent structures that did not interact with the adjacent protein domains. Their length, composition, hydrophobicity, and secondary structure together made important contributions towards achieving the desirable functions. Natural linkers could serve as a general reference for the rational design of empirical linkers in recombinant fusion proteins.
Empirical linkers in recombinant fusion proteins
In addition to the many candidate linkers identified from studies on naturally occurring protein linkers, scientists have also designed empirical linkers with a variety of sequences and confirmations for recombinant fusion protein production. These empirical linkers are broadly classified into: flexible linkers, rigid linkers and cleavable linkers.
- Flexible linkers: Flexible linkers are usually applied when the protein domains that need to be joined require a certain degree of movement or interaction. They are generally composed of small, non-polar (e.g. Gly) or polar (e.g. Ser or Thr) amino acids. The small size of these amino acids provides flexibility, and allows for mobility of the connecting functional domains. The incorporation of Ser or Thr can maintain the stability of the linker in aqueous solutions by forming hydrogen bonds with the water molecules, and therefore reduce the unfavorable interaction between the linker and the protein moieties. An example of the most widely used flexible linker is the sequence (Gly-Gly-Gly-Gly-Ser)n.
- Rigid linkers: While flexible linkers have the advantage of connecting the functional domains passively and permitting a certain degree of movement, the lack of rigidity of these linkers can be a limitation. There are several examples in the literature where the use of flexible linkers resulted in poor expression yields or loss of biological activity. For instance, a Tf-granulocyte colony stimulating factor (G-CSF) fusion protein failed to be expressed with a flexible (GGGGS)3 linker. In another report, the immunoglobulin binding ability of the protein G domain in a protein G-Vargula luciferase fusion protein was not recovered after inserting a flexible GGGGS linker. The ineffectiveness of flexible linkers in these instances was attributed to an inefficient separation of the protein domains or insufficient reduction of their interference with each other. Under these situations, rigid linkers have been successfully applied to keep a fixed distance between the domains and to maintain their independent functions. Rigid linkers exhibit relatively stiff structures by adopting α-helical conformations or by containing multiple Pro residues. Examples of some rigid linkers are: (EAAAK)n and (XP)n, with X designating any amino acid, preferably Ala, Lys, or Glu.
- In vivo cleavable linkers: Flexible and rigid linkers represent stable linkers that covalently join functional protein domains together to act as one molecule throughout the in vivo processes that the component protein(s) are involved in. This stable linkage between functional domains provides many advantages such as a prolonged plasma half-life (e.g. albumin or Fc-fusions).
However, it also has several potential drawbacks including steric hindrance between functional domains, decreased bioactivity, and altered biodistribution and metabolism of the protein moieties due to the interference between domains. Under such circumstances, cleavable linkers are used to release free functional domains in vivo. This type of linker may reduce steric hindrance, improve bioactivity, or achieve independent actions/metabolism of individual domains of recombinant fusion proteins after linker cleavage. The design of in vivo cleavable linkers in recombinant fusion proteins is quite challenging. Unlike the versatility of crosslinking agents available for chemical conjugation methods, linkers in recombinant fusion proteins must necessarily be oligopeptides. Some examples of in vivo cleavable linkers are described below:
An in vivo cleavable disulfide linker (LEAGCKNFFPR↓SFTSCGSLE), based on the reversible nature of the disulfide bond, was designed for recombinant fusion proteins by Chen et al. (3), and offered the advantage of generating a precisely constructed, homogeneous product by recombinant methods. This disulfide linker was based on a dithiocyclopeptide containing an intramolecular disulfide bond formed between two cysteine (Cys) residues on the linker, as well as a thrombin-sensitive sequence (PRS) between the two Cys residues (Figure 1). This linker was inserted between G-CSF and Tf to construct a model fusion protein (designated as “G-C-T”). The in vitro thrombin treatment of G-C-T resulted in the cleavage of the thrombin-sensitive sequence, while the reversible disulfide linkage between the two domains of the fusion protein remained. The resultant disulfide-linked protein was designated as “G- S-S-T”. This disulfide-linked fusion protein was demonstrated to be cleavable in vivo following intravenous administration to CF1 mice. A rapid release of G-CSF from G-S-S-T in the blood was observed as early as 5 minutes, with a peak at ~15 minutes post injection. The released free G-CSF exhibited a quick elimination due to its short in vivo half-life. In contrast, no detectable amount of free G-CSF was released in vivo from G-C-T, which has a stable peptide linker. This study demonstrated the construction of a disulfide-linked fusion protein for use in applications where the in vivo separation of protein domains is desired.
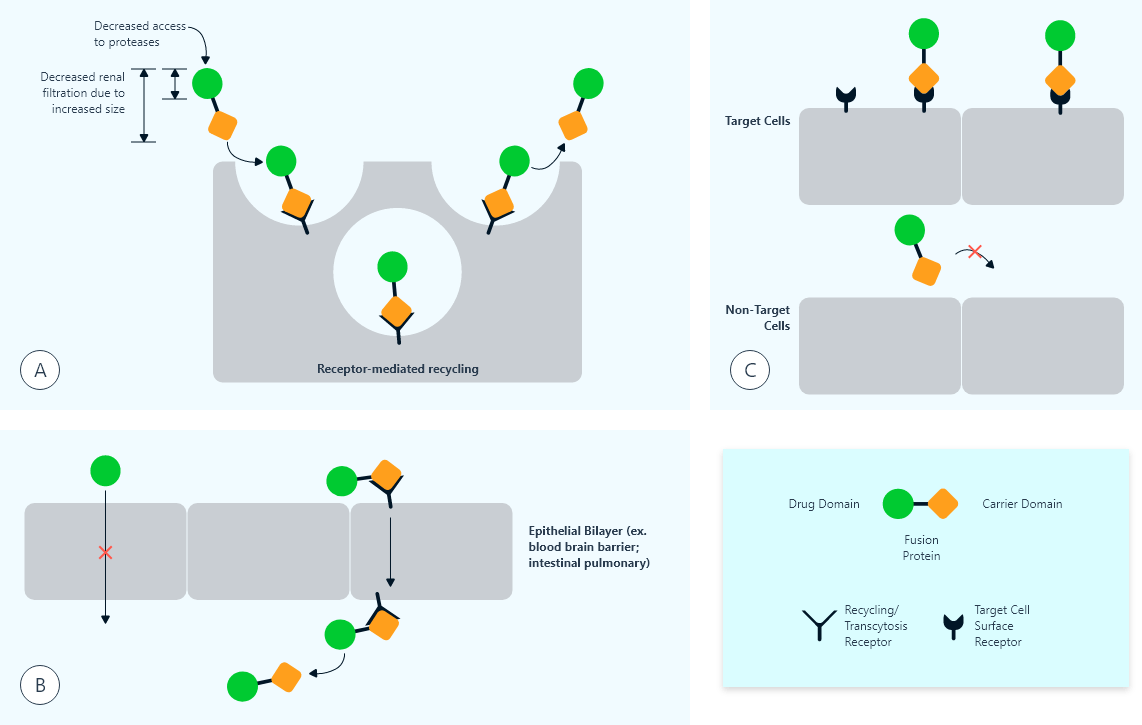
Fig.1: Illustration of in vivo cleavable linkers
More recently, a similar cyclopeptide linker was designed to create an in vivo cleavable disulfide linker in an interferon-α2b (IFN-α2b) and HSA fusion protein (4). The dithiocyclopeptide sequence (CRRRRRREAEAC) contains an intramolecular disulfide bond between 2 Cys residues, as well as a peptide sequence sensitive to the secretion signal processing proteases resident in the yeast secretory pathway. During the protein expression, the linker was first cleaved by protease Kex2 at CRRRRRR↓EAEAC, followed by cleavage of proteases Kex1 and Ste13. As a result, the amino acids between two Cys residues in the linker were completely removed during secretion, and the disulfide linked fusion protein was directly expressed from Pichia pastoris.
The in vivo cleavage of the linkers in recombinant fusion proteins may also be carried out by proteases that are expressed in vivo under pathological conditions (e.g. cancer or inflammation), in specific cells or tissues, or constrained within certain cellular compartments (Figure 1). Such in vivo cleavable linkers are designed to incorporate specific protease-sensitive sequences. Unlike the reduction of disulfide bonds which happens rapidly in the blood circulation (3), the specificity of many proteases offers slower cleavage of the linker in constrained compartments. Thus, this type of cleavable linker can be applied to activation of fusion protein bioactivity at specific sites in vivo.
Overall, linkers can adopt various structures and exert diverse functions to fulfill the application of fusion proteins (Table 2). The flexible linkers are often rich in small or hydrophilic amino acids such as Gly or Ser to provide structural flexibility and have been applied to connect functional domains that favor interdomain interactions or movements. In cases where sufficient separation of protein domains is required, rigid linkers may be preferable. By adopting α-helical structures or incorporating Pro, the rigid linkers can efficiently keep protein moieties at a distance. Both flexible and rigid linkers are stable in vivo, and do not allow the separation of joined proteins. Cleavable linkers, on the other hand, permit the release of free functional domains in vivo via reduction or proteolytic cleavage. They can be utilized to improve the bioactivity of chimeric proteins, or to specifically deliver prodrugs to target sites where the linkers are processed to activate bioactivity. The rational choice of linkers should be based on the properties of the linkers and the desired fusion proteins.
Table 2: Summary of empirical linkers
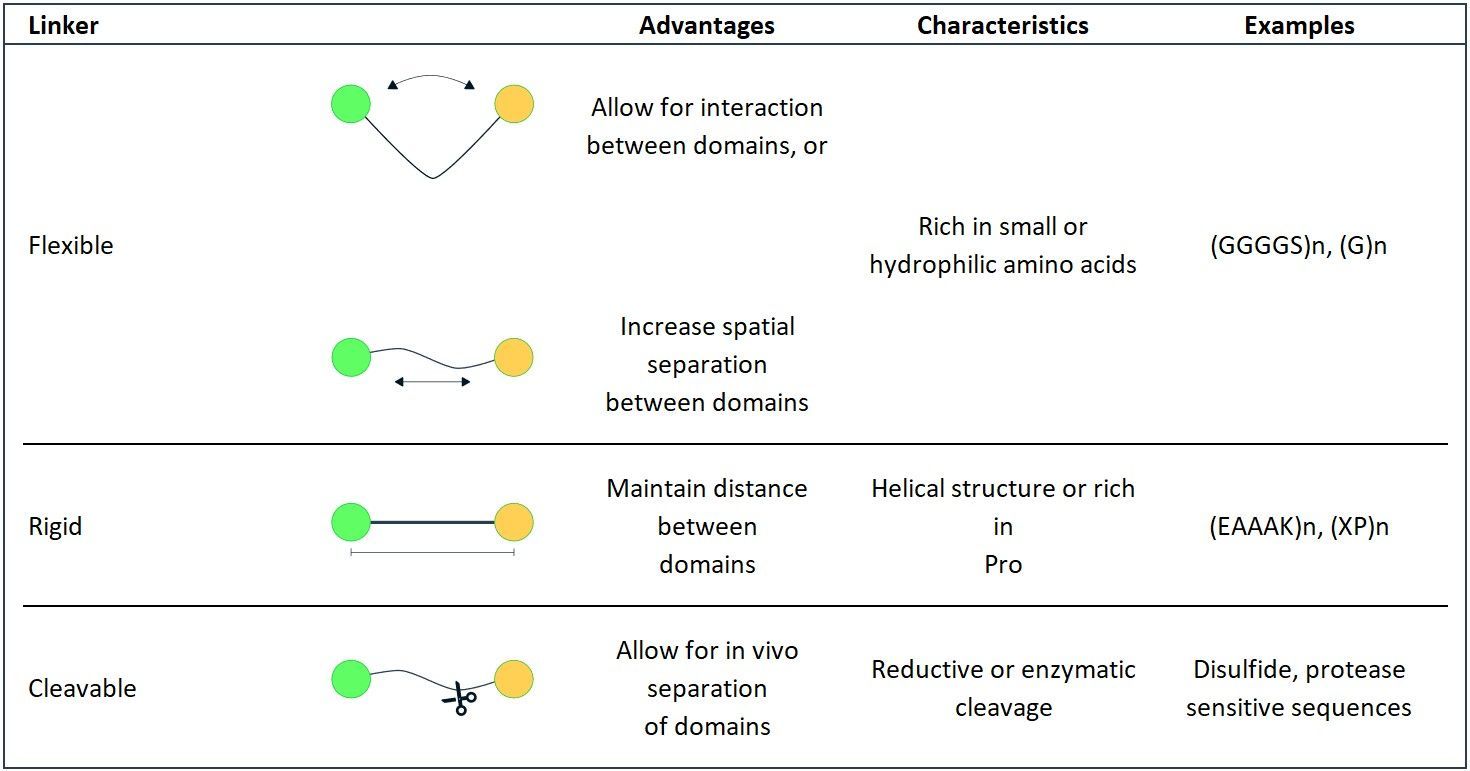
Table 2: Summary of empirical linkers
Functionality of linkers in fusion proteins
Apart from their most basic function of covalently joining functional domains of proteins and release them under desired conditions, they also contribute to derived functions such as improving expression yields, biological activity and stability. Some important additional functions of linkers are described below-
Linkers can improve folding and stability of fusion proteins:
The flexible GS linker has been shown to improve folding and stability in several fusion proteins. A very important application of the flexible GS linker is the construction of single-chain variable fragment (scFv), an antigen-binding fusion protein composed of antibody light-chain variable region (VL) tethered to heavy chain variable region (VH) via an oligopeptide linker (5). A flexible linker (GGGGS)3 was designed by Huston et al. to construct a scFv, since its flexible structure would allow for the correct orientation of the VH and VL domains, and would not interfere with the folding of the protein domains (6). The length of the linker was adjusted according to the distance between the C-terminus of the VH domain and the N-terminus of the VL domain under its natural orientation (3.5 nm). The length of the (GGGGS)3 linker was calculated to be about 5.7 nm, and was expected to bridge the VH and VL domains (6). This (GGGGS)3 linker was shown to be suitable for constructing scFv due to its high flexibility, and has since been applied to many other scFvs (7-9). In addition to flexible linkers, helical linkers can also improve fusion protein folding and stability.
Linkers can improve expression of fusion proteins:
Besides impaired biological activity, the difficulty to express stable and high levels of recombinant fusion protein is often another hurdle encountered during the application of fusion proteins for drug delivery. Due to the structural perturbation between protein domains, fusion proteins may be misfolded, unstable and appear as a heterogeneous product, often resulting in a low expression yield. Although the expression of fusion proteins can sometimes be improved by simply switching the orientation of the component protein domains, the interference may not be effectively reduced due to the short distance between domains. Since many linkers can keep domains at proper distance and allow for their independent folding, they can serve as practical tools to enhance the expression yield of recombinant fusion proteins.
Linkers can improve bioactivity of fusion proteins:
By fusing two or more protein domains, a fusion protein usually acquires the biological activities from each component. However, the direct fusion of proteins often results in impaired biological activity, probably because the functional domains are brought too close to properly interact with their corresponding binding proteins (i.e. receptors or ligands). Under these circumstances, linkers may be very effective tools to maintain an appropriate distance between domains to reduce their interference, restore or improve folding, or allow for the in vivo release of the free protein drug domain to ultimately improve bioactivity. The bioactivity of fusion proteins can also be improved by adjusting the length of linkers to increase the space between component proteins.
Linkers can target fusion proteins to specific sites in vivo:
Linker insertion between fusion protein domains can also improve or enable targeting of fusion protein to specific sites in vivo. One way in which linkers can improve targeting is simply by increasing the binding affinity of the targeting protein domain for its receptor. Linkers can provide distance between domains, reduce their interference, and ultimately improve their receptor binding affinity. A second approach for application of linkers to improve drug targeting involves introduction of a linker sequence that will enable specific activation of the fusion protein at the target site. In this approach, the intact fusion protein shows reduced or no biological activity, but the cleavage of the linker at specific sites releases the free, biologically active protein drug domain at the target site (Figure 2).
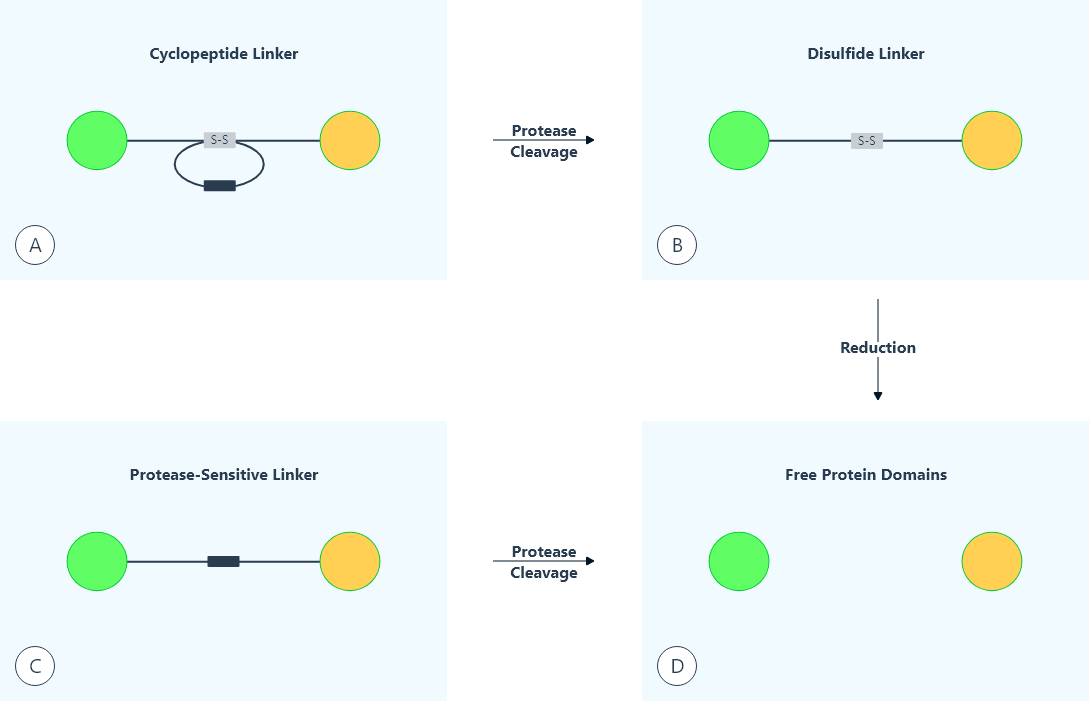
Fig. 2: Use of linkers to target fusion proteins to specific sites in vivo
For example, a protease present in the lysosome, cathepsin B, has been applied for targeted intracellular activation of cytotoxic proteins. Cathepsin B substrate peptides have previously been utilized as cleavable peptide linkers in many bioconjugates (10, 11). For instance, a dipeptide of Phe-Lys was applied to serve as part of a cleavable linker in an albumin-binding prodrug of doxorubicin 1, for the in vivo release of doxorubicin after Cathepsin B cleavage in the tumor. The cathepsin B-cleavable linkers have recently been applied to fusion proteins. Yuan et. al. used a cathepsin B sensitive peptide of GFLG together with a furin cleavage sequence of R2KR6, to link a tumor-targeting moiety (fragment of C. perfringens enterotoxin) and a toxin (recombinant gelonin) in order to release the toxin in the lysosome (12).
Linkers can affect the PK of fusion proteins:
Fusion proteins provide many advantages over the parent proteins, such as improved PK and PD properties as in albumin- and Fc-fusion proteins, as well as the drug targeting effects seen in immunotoxins. Although several fusion proteins have been applied in the clinic, the mechanisms underlying PK of bifunctional fusion proteins are still largely unexplored, and a generalized PK model for fusion proteins is not established. Target-mediated drug disposition (TMDD), which describes the process where drug-target binding significantly influences the PK and PD of the drug, has been established as a crucial mechanism for the elimination of many single domain protein and peptide drugs (14). Generally, for many protein drugs, the disposition processes affecting their PK are relatively simple, e.g., binding to their cell surface receptor leads to endocytosis and lysosomal degradation. However, the disposition of bifunctional fusion proteins are affected by two different domains/binding sites, and therefore their PK/PD properties are much more complicated. Since linker insertion may alter the receptor binding affinity of each protein domain, it can affect the in vivo disposition of fusion proteins and increase the complexity of PK studies.
Summary
During the development of therapeutic recombinant fusion proteins, linker design has become a valuable means to achieve desired characteristics of the products. Linker sequences derived from natural multi-domain proteins may provide useful references for designing empirical linkers. Various empirical linkers such as flexible, rigid or cleavable linkers have been designed for various purposes, such as passively joining domains, spatially separating domains, or releasing free functional domains in vivo. Optimal linkers can provide many advantages for the production of fusion proteins, including improving structural stability, enhancing bioactivity, increasing expression levels, altering the PK profiles and enabling the in vivo targeting of the fusion proteins. Although many examples of various types of linkers have been developed in the past, the rational design of linkers for the construction of fusion proteins is still in its infancy. Systematic, strategic scientific endeavors are in demand to greatly advance the science of linker design and application. Many technology platforms may be investigated in more depth towards understanding the connection between linker composition and structure, and ultimately tie them to linker function.
The study of linker composition and structure, and the investigation of linker function should go hand in hand when designing a novel linker. A good example of the rational design of linkers are the rigid helical linkers (A(EAAAK)nA) by Arai et al. (15, 16). The idea of using these sequences as a linker developed from the finding that they form an α-helical conformation in water as determined by circular dichroism (17). It was then proposed to apply them to effectively separate protein domains in fusion proteins.
Another fruitful direction would be the establishment of more databases and searching programs for linkers. With the rapid advancement of protein science and biotechnology, the design of linkers in fusion proteins has become more important than ever before. With a thorough understanding of their structures, conformations, and functions via future biomedical research, the incorporation of linkers will greatly facilitate the construction of stable and bioactive recombinant fusion proteins for drug delivery applications.
Get Your Free QUICK-LINKS Sheet
References
- Argos P. An investigation of oligopeptides linking domains in protein tertiary structures and possible candidates for general gene fusion. J Mol Biol. 1990; 211:943–958.
- George R, Heringa J. An analysis of protein domain linkers: their classification and role in protein folding. Protein Eng. 2002; 15:871–879.
- Chen X, Bai Y, Zaro J, Shen WC. Design of an in vivo cleavable disulfide linker in recombinant fusion proteins. Biotechniques. 2010; 49:513–518.
- Zhao HL, Xue C, Du JL, Ren M, Xia S, Liu ZM. Balancing the pharmacokinetics and pharmacodynamics of interferon-alpha2b and human serum albumin fusion protein by proteolytic or reductive cleavage increases its in vivo therapeutic efficacy. Mol Pharm. 2012; 9:664–670.
- Hagemeyer CE, von Zur Muhlen C, von Elverfeldt D, Peter K. Single-chain antibodies as diagnostic tools and therapeutic agents. Thromb Haemost. 2009; 101:1012–1019.
- Huston J, Levinson D, Mudgett-Hunter M, Tai M, Novotný J, Margolies M, Ridge R, Bruccoleri R, Haber E, Crea R. Protein engineering of antibody binding sites: recovery of specific activity in an anti-digoxin single-chain Fv analogue produced in Escherichia coli. Proc Natl Acad Sci U S A. 1988; 85:5879–5883.N
- Trinh R, Gurbaxani B, Morrison SL, Seyfzadeh M. Optimization of codon pair use within the (GGGGS)3 linker sequence results in enhanced protein expression. Mol Immunol. 2004; 40:717– 722.
- Reddy ST, Ge X, Miklos AE, Hughes RA, Kang SH, Hoi KH, Chrysostomou C, Hunicke-Smith SP, Iverson BL, Tucker PW, Ellington AD, Georgiou G. Monoclonal antibodies isolated without screening by analyzing the variable-gene repertoire of plasma cells. Nat Biotechnol. 2010; 28:965–969.
- Alfthan K, Takkinen K, Sizmann D, Söderlund H, Teeri TT. Properties of a single-chain antibody containing different linker peptides. Protein Eng. 1995; 8:725–731.e
- Peterson JJ, Meares CF. Cathepsin substrates as cleavable peptide linkers in bioconjugates, selected from a fluorescence quench combinatorial library. Bioconjug Chem. 1998; 9:618–626.
- Abu Ajaj K, Graeser R, Fichtner I, Kratz F. In vitro and in vivo study of an albumin-binding prodrug of doxorubicin that is cleaved by cathepsin B. Cancer Chemother Pharmacol. 2009; 64:413–418.
- Yuan X, Lin X, Manorek G, Howell SB. Challenges associated with the targeted delivery of gelonin to claudin-expressing cancer cells with the use of activatable cell penetrating peptides to enhance potency. BMC Cancer. 2011; 11:61.
- Mager DE. Target-mediated drug disposition and dynamics. Biochem Pharmacol. 2006; 72:1–10.
- Mager DE. Target-mediated drug disposition and dynamics. Biochem Pharmacol. 2006; 72:1–10.
- Arai R, Ueda H, Kitayama A, Kamiya N, Nagamune T. Design of the linkers which effectively separate domains of a bifunctional fusion protein. Protein Eng. 2001; 14:529–532.
- Arai R, Wriggers W, Nishikawa Y, Nagamune T, Fujisawa T. Conformations of variably linked chimeric proteins evaluated by synchrotron X-ray small-angle scattering. Proteins. 2004; 57:829– 838.
- Marqusee S, Baldwin RL. Helix stabilization by Glu-...Lys+ salt bridges in short peptides of de novo design. Proc Natl Acad Sci U S A. 1987; 84:8898–8902.
- Chen X, Zaro JL, Shen WC. Fusion protein linkers: property, design and functionality. Adv Drug Deliv. Rev. 2013;65(10):1357-1369.