Application of Covalent Bonds Between Small Molecules and Proteins for Improved Drug Discovery
Application of Covalent Bonds Between Small Molecules and Proteins for Improved Drug Discovery
Insight on current warhead modalities and the potential of covalence chemistry earlier in the target discovery process
Introduction
Covalent bonds play a key role in guiding the mechanisms of biology. Along with binding nucleic acids, amino acids, protein and the other biomolecules of life; covalence principles are rooted into all the drugs developed in today’s market. Although their dependency is high, our capabilities in manipulating covalent bond-forming reactions to correlating biofunction remains restricted. This impediment demonstrates areas for improvements in our discovery approaches and platform technologies.
For example, the conventional methods for identifying novel hits involves a series of technologies - built around: high-throughput or phenotypic screening, fragment-specific analysis, and DNA library parsing. All which utilize chemical matter that binds to targets of interest via non-covalent interactions while neglecting covalent factors. Reason being that these systems are not fit to handle the attrition caused by reactive functionalities. Therefore, covalent compounds are often disregarded from these approaches. In recent years the successes of targeting covalent inhibitors has increased the consideration of covalent molecules in drug discovery. This revolution is due to the improved understanding of the factors governing the binding of covalent compounds and it has generated a growing focus on enhancing the non-covalent recognition to biological targets of interest – along with reducing off-targeting risks. There are other reports available that do a much better job reviewing these concepts and platforms for drug discovery. In this piece, we will focus on surveying select examples of electrophile modalities advancing the small molecule chemistry in drug discovery. 1-17
Common nucleophile warheads
Covalent bond formation is a result of an electrophile reacting with a nucleophile. The preferred nucleophile has traditionally been a cysteine thiol in biological systems because its high reactivity and low prevalence in the proteome – making it a low risk candidate for random reactions. Cysteine-based covalent chemistry has been very successful overall in delivering safe and specific drugs. This has made it an essential tool in the drug hunters toolbox against challenging targets. 12,13,18-25
Beyond the cysteine-based domain, novel nucleophile to electrophile pairs are breaking ground throughout scientific literature. One amino acid that has been rising in fame is lysine. Various papers are detailing the effectiveness of lysine-reactive warheads. Particularly in; activated esters, sulfur-based reactive centers and α, β-unsaturated modalities. While featuring high stability and selectivity, lysine-reactive warheads present notable advantages than structurally similar reversible inhibitors. 26-31
Increasingly novel modalities
Serine is one of a few alcoholic side chains that can be targeted via multiple electrophile warheads including activated esters, fluorophosphates and sulfur centers. While histidine has been proven to react with α, β -unsaturated systems, epoxides, phosphorus reagents and sulfonyl fluorides. 34-40
With minimal success, negatively charged side chains such as aspartic acid and glutamic acid are rare considerations in this area. However, they should not be completely disregarded since there is evidence to support their application as warheads with Woodward’s reagent K. 54
Contrary in its properties, recent success with methionine in hypervalent iodine reagents and redox-based systems earns it an honorable mention. A more novel modality, methionine presents an unattractive characteristic of low nucleophilicity. However, covalent modifications of its residues have suggested promising solutions in countering such negative features along with better compliance towards native proteomics. Which in turn hints that both the application of modifying covalent residues and methionine as a modality will likely pick up traction and popularity in future warhead concept designs. 55-57, 68
To help visualize all the electrophiles we explained, we generated a corresponding figure chart below.
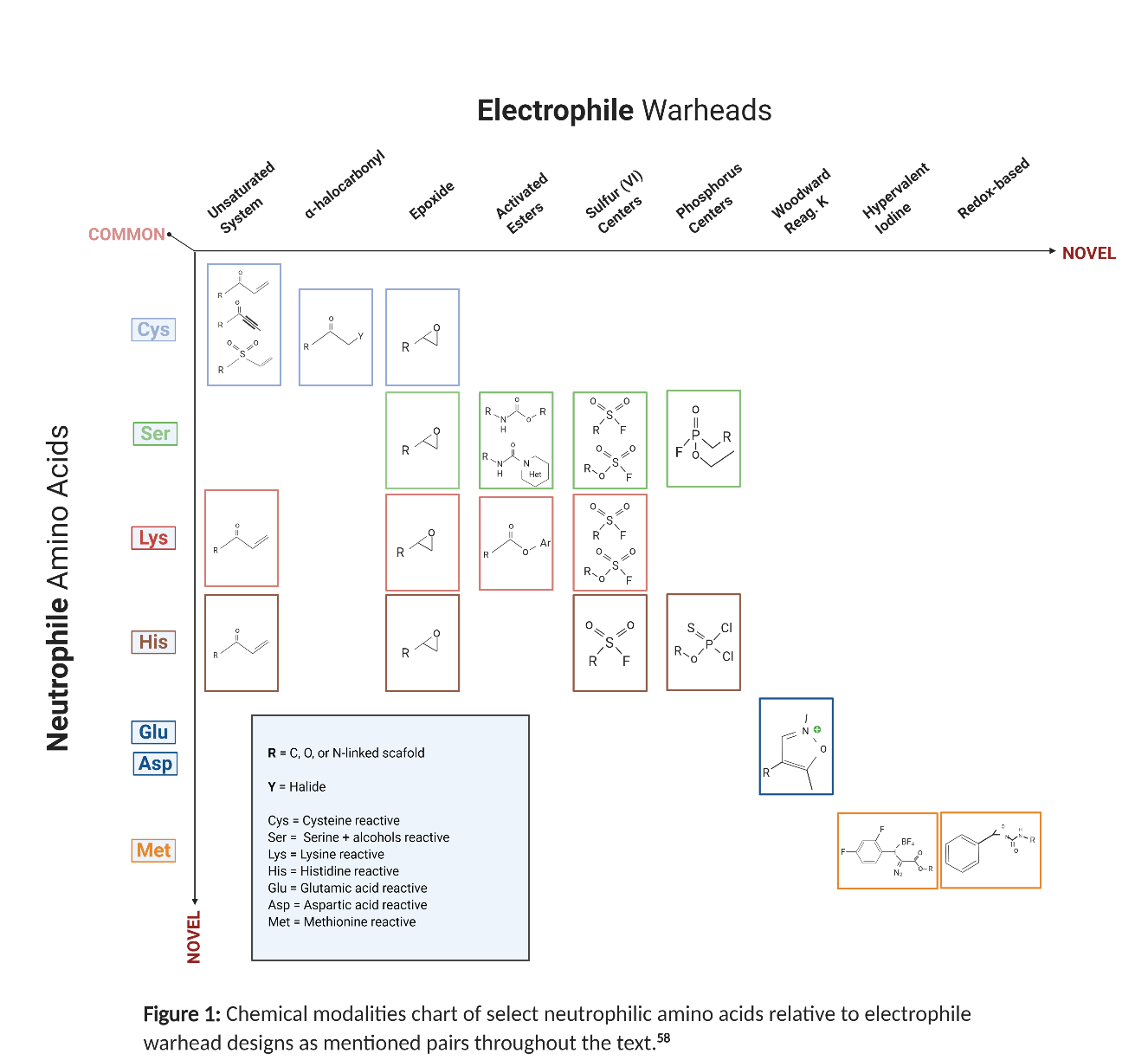
Fig 1: Chemical modalities chart of select neutrophilic amino acides relative to electrophile warhead designs as mentioned pairs throughout the text. 58
A lot of these electrophiles involve more complex factors and information beyond what is presented. The list of references provided at the end is our recommended source for the appropriate comprehensive information.
Chemical Criteria
Warheads are complicated and the potential for new or mixed pairs are endless. Ultimately, this comes down to three major principles in design. First, the electrophiles need to meet a minimum reactivity threshold, demonstrating low risk for promiscuous reactivity and off-target binding. This can be determined by testing the chemical reaction rate against a model nucleophile or analogue. Second, it must be optimizable so that the warhead can be positioned in favor of the target residue. This is critical in achieving efficiency between the covalent reaction and the desired targets (Kinact). Optimizing this type of structural confirmation can be supported by traditional medicinal chemistry techniques, guided by the required chemical mechanism for achieving a covalent reaction (i.e. sigma, Pi bond, orbital overlap). Finally, a reactive functional group must be available on the target protein in order for any of the warhead chemistry to be achieved.
Functional Target
The principle of targeting an appropriate functional group on a protein mainly applies when developing covalent drugs in a clinical application. This is an important note because unlike in clinical applications, non-clinical settings provide the utility of external factors which can support guiding target function. So, factors such as light can be harnessed in this sense. One popular example for light is the photoaffinity labelling (PAL) method. PAL utilizes probes that contain photoactivate functions and binds them to nearby bonds via UV irradiation activation. This method has helped develop several probes and is frequently used to target novel binding pockets in proteins (that may be absent nucleophilic amino acids). 9,38,60-63
Along with binding support, PAL is one of several applications useful for identifying specifics of an interaction that go beyond time-dependent techniques. Employing these applications with conventional methods shows great promise in providing the missing pieces for characterizing small molecules for drug discovery platforms. Particularly when it comes to target engagement/validation, selectivity profiling and covalent frameworks for protein modification. 64-67
By providing this overview of the relationship between chemical warhead modalities and covalent bonding, we will be able to follow up with reviews expanding on the individual application technologies mentioned such as PAL.
References
- J. R. Broach, J. Thorner, in Nature, Elsevier Inc., 1996, pp. 14–16.
- C. W. Murray, D. C. Rees, Nat. Chem. 2009, 1, 187–192.
- D. E. Scott, A. G. Coyne, S. A. Hudson, C. Abell, Biochemistry 2012, 51, 4990–5003.
- R. A. Goodnow, C. P. Davie, in Annu. Rep. Med. Chem., Elsevier Inc., 2017, pp. 1–15.
- J. G. Moffat, F. Vincent, J. A. Lee, J. Eder, M. Prunotto, Nat. Rev. Drug Discov.2017, 16, 531–543.
- M. H. Potashman, M. E. Duggan, J. Med. Chem. 2009, 52, 1231–1246.
- B. K. Park, A. Boobis, S. Clarke, C. E. P. Goldring, D. Jones, J. G. Kenna, C. Lambert, H. G. Laverty, D. J. Naisbitt, S. Nelson, et al., Nat. Rev. Drug Discov. 2011, 10, 292–306.
- S. De Cesco, J. Kurian, C. Dufresne, A. K. Mittermaier, N. Moitessier, Eur. J. Med. Chem. 2017, 138, 96–114.
- J. Singh, R. C. Petter, T. A. Baillie, A. Whitty, Nat. Rev. Drug Discov. 2011, 10, 307–317.
- R. A. Bauer, Drug Discov. Today 2015, 20, 1061–1073.
- A. A. Adeniyi, R. Muthusamy, M. E. Soliman, Expert Opin. Drug Discov. 2016, 11, 79–90.
- T. A. Baillie, Angew. Rev. 2016, 55, 2–17.
- Z. Zhao, P. E. Bourne, Drug Discov. Today 2018, 23, 727–735.
- R. Lonsdale, R. A. Ward, Chem. Soc. Rev. 2018, 47, 3816–3830.
- A. Tuley, W. Fast, Biochemistry 2018, 57, 3326–3337.
- T. Zhang, J. M. Hatcher, M. Teng, N. S. Gray, M. Kostic, Cell Chem. Biol. 2019, DOI 10.1016/j.chembiol.2019.09.012.
- S. Ray, A. S. Murkin, Biochemistry 2019, DOI 10.1021/acs.biochem.9b00293.
- S. M. Marino, V. N. Gladyshev, J. Mol. Biol. 2010, 404, 902–916.
- Z. Zhao, Q. Liu, S. Bliven, L. Xie, P. E. Bourne, J. Med. Chem. 2017, 60, 2879– 2889.
- M. Visscher, M. R. Arkin, T. B. Dansen, Curr. Opin. Chem. Biol. 2016, 30, 61–67.
- Q. Liu, Y. Sabnis, Z. Zhao, T. Zhang, S. J. Buhrlage, L. H. Jones, N. S. Gray, Chem. Biol. 2013, 20, 146–159.
- M. Gehringer, S. A. Laufer, J. Med. Chem. 2019, 62, 5673–5724.
- R. Lonsdale, J. Burgess, N. Colclough, N. L. Davies, E. M. Lenz, A. L. Orton, R. A. Ward, J. Chem. Inf. Model. 2017, 57, 3124–3137.
- L. Garuti, M. Roberti, G. Bottegoni, Curr. Med. Chem. 2011, 18, 2981–2994.
- H. Cheng, S. Planken, ACS Med. Chem. Lett. 2018, 9, 861–863.
- H. Mukherjee, N. P. Grimster, Curr. Opin. Chem. Biol. 2018, 44, 30–38.
- E. Mons, I. D. C. Jansen, J. Loboda, B. R. van Doodewaerd, J. Hermans, M. Verdoes, C. A. A. van Boeckel, P. A. van Veelen, B. Turk, D. Turk, et al., J. Am. Chem. Soc. 2019, 141, 3507–3514.
- M. E. Schnute, S. E. Benoit, I. P. Buchler, N. Caspers, M. L. Grapperhaus, S. Han, R. Hotchandani, N. Huang, R. O. Hughes, B. M. Juba, et al., ACS Med. Chem. Lett. 2019, 10, 80–85.
- J. M. Bradshaw, J. M. McFarland, V. O. Paavilainen, A. Bisconte, D. Tam, V. T. Phan, S. Romanov, D. Finkle, J. Shu, V. Patel, et al., Nat. Chem. Biol. 2015, 11,525–531.
- A. Casimiro-Garcia, J. I. Trujillo, F. Vajdos, B. Juba, M. E. Banker, A. Aulabaugh, P. Balbo, J. Bauman, J. Chrencik, J. W. Coe, et al., J. Med. Chem. 2018, 61, 10665–10699.
- J. Pettinger, K. Jones, M. D. Cheeseman, Angew. Chemie Int. Ed. 2017, 56, 15200–15209.
- S. E. Dalton, L. Dittus, D. A. Thomas, M. A. Convery, J. Nunes, J. T. Bush, J. P. Evans, T. Werner, M. Bantscheff, J. A. Murphy, et al., J. Am. Chem. Soc. 2018, 140, 932–939.
- S. Choi, S. Connelly, N. Reixach, I. A. Wilson, J. W. Kelly, Nat. Chem. Biol. 2010, 6, 133–139.
- N. P. Grimster, S. Connelly, A. Baranczak, J. Dong, L. B. Krasnova, K. B. Sharpless, E. T. Powers, I. A. Wilson, J. W. Kelly, J. Am. Chem. Soc. 2013, 135, 5656–5668.
- N. N. Gushwa, S. Kang, J. Chen, J. Taunton, J. Am. Chem. Soc. 2012, 134, 20214–20217.
- L. H. Jones, ACS Med. Chem. Lett. 2018, 9, 584–586.
- N. Wang, B. Yang, C. Fu, H. Zhu, F. Zheng, T. Kobayashi, J. Liu, S. Li, C. Ma, P. G. Wang, et al., J. Am. Chem. Soc. 2018, 140, 4995–4999.
- U. Dahal, A. Gilbert, R. Obach, J. Chen, C. Garcia-Irizarry, B. Schuff, J. Starr, D. Uccello, J. Young, Med. Chem. Commun. 2016, 7, 864–872.
- E. Anscombe, E. Meschini, R. Mora-vidal, R. Stephen, J. A. Endicott, R. J. Griffin, Chem. Biol. 2015, 22, 1159–1164.
- J. Pettinger, Y.-V. Le Bihan, M. Widya, R. L. M. Van Montfort, K. Jones, M. D. Cheeseman, Angew. Chemie Int. Ed. 2017, 56, 3536–3540.
- H. Mukherjee, J. Debreczeni, J. Breed, S. Tentarelli, B. Aquila, J. E. Dowling, A. Whitty, N. P. Grimster, Org. Biomol. Chem. 2017, 15, 9685–9695.
- C. Baggio, L. Gambini, P. Udompholkul, A. F. Salem, A. Aronson, A. Dona, E. Troadec, F. Pichiorri, M. Pellecchia, J. Med. Chem. 2018, 61, 6350–6363.
- Y. Liu, M. P. Patricelli, B. F. Cravatt, Proc. Natl. Acad. Sci. 1999, 96, 14694– 14699.
- A. Narayanan, L. H. Jones, Chem. Sci. 2015, 6, 2650–2659.
- K. Ahn, D. S. Johnson, M. Mileni, D. Beidler, J. Z. Long, M. K. McKinney, E. Weerapana, N. Sadagopan, M. Liimatta, S. E. Smith, et al., Chem. Biol. 2009, 16, 411–420.
- A. B. Cognetta, M. J. Niphakis, H.-C. Lee, M. L. Martini, J. J. Hulce, B. F.Cravatt, Chem. Biol. 2015, 22, 928–937.
- J. W. Chang, M. J. Niphakis, K. M. Lum, A. B. Cognetta, C. Wang, M. L. Matthews, S. Niessen, M. W. Buczynski, L. H. Parsons, B. F. Cravatt, Chem. Biol. 2012, 19, 579–588.
- J. S. Cisar, O. D. Weber, J. R. Clapper, J. L. Blankman, C. L. Henry, G. M. Simon, J. P. Alexander, T. K. Jones, R. A. B. Ezekowitz, G. P. O’Neill, et al., J. Med. Chem. 2018, 61, 9062–9084.
- S. Liu, Sci. 1998, 282, 1324–1327.
- D. A. Bullough, W. S. Allison, J. Biol. Chem. 1986, 261, 5722–5730.
- M. Yoshizawa, T. Itoh, T. Hori, A. Kato, Y. Anami, N. Yoshimoto, K. Yamamoto, J. Med. Chem. 2018, 61, 6339–6349.
- A. de Saint Germain, G. Clavé, M.-A. Badet-Denisot, J.-P. Pillot, D. Cornu, J.-P. Le Caer, M. Burger, F. Pelissier, P. Retailleau, C. Turnbull, et al., Nat. Chem. Biol. 2016, 12, 787–794.
- S. Jia, D. He, C. J. Chang, J. Am. Chem. Soc. 2019, 141, 7294–7301.
- P. Martín-Gago, E. K. Fansa, M. Winzker, S. Murarka, P. Janning, C. Schultz- Fademrecht, M. Baumann, A. Wittinghofer, H. Waldmann, Cell Chem. Biol.2017, 24, 589–597.
- S. Lin, X. Yang, S. Jia, A. M. Weeks, M. Hornsby, P. S. Lee, R. V. Nichiporuk, A. T. Iavarone, J. A. Wells, F. D. Toste, et al., Science 2017, 355, 597–602.
- L. B. Poole, Free Radic. Biol. Med. 2015, 80, 148–157.
- M. T. Taylor, J. E. Nelson, M. G. Suero, M. J. Gaunt, Nature 2018, 562, 563– 568.
- Dalton, S.E. and Campos, S. (2020), Covalent Small Molecules as Enabling Platforms for Drug Discovery. ChemBioChem. doi:10.1002/cbic.201900674
- O. A. Kharenko, R. G. Patel, S. D. Brown, C. Calosing, A. White, D. Lakshminarasimhan, R. K. Suto, B. C. Duffy, D. B. Kitchen, K. G. McLure, et al., J. Med. Chem. 2018, 61, 8202–8211.
- V. J. Cee, L. P. Volak, Y. Chen, M. D. Bartberger, C. Tegley, T. Arvedson, J. McCarter, A. S. Tasker, C. Fotsch, J. Med. Chem. 2015, 58, 9171–9178.
- J. S. Martin, C. J. MacKenzie, D. Fletcher, I. H. Gilbert, Bioorg. Med. Chem. 2019, 27, 2066–2074.
- R. A. Ward, M. J. Anderton, S. Ashton, P. A. Bethel, M. Box, S. Butterworth, N. Colclough, C. G. Chorley, C. Chuaqui, D. A. E. Cross, et al., J. Med. Chem. 2013, 56, 7025–7048.
- J. M. Strelow, SLAS Discov. 2017, 22, 3–20.
- A. Singh, E. R. Thornton, F. H. Westheimer, J. Biol. Chem. 1962, 237, PC3006–PC3008.
- L. Dubinsky, B. P. Krom, M. M. Meijler, Bioorganic Med. Chem. 2012, 20, 554–570.
- E. Smith, I. Collins, Future Med. Chem. 2015, 7, 159–183.
- P. P. Geurink, L. M. Prely, G. A. van der Marel, R. Bischoff, H. S. Overkleeft, in Act. Protein Profiling. Top. Curr. Chem. (Ed.: S.L. Schreiber), Springer- Verlag, Berlin, Heidelberg, 2011, pp. 85–113.
- J. Zang, Y. Chen, W. Zhu, S. Lin, Biochemistry 2019, acs.biochem.9b00789.