Viral RNA Isolation Methods Reviewed: Spin vs. Magnetic
Viral RNA Isolation Methods Reviewed: Spin vs. Magnetic
Shalaka Samant, Ed Hamdeh, Cindy Lee
kbDNA INC. – Cambridge, MA U.S.A
Introduction
Nucleic acid extraction is the primary step for many downstream applications in molecular biology. Genomic (or chromosomal) DNA, plasmids, and different types of RNAs represent the broad categories of intracellular nucleic acids that are typically isolated from a variety of samples. High-quality yields of contamination-free RNA are required for downstream use in various applications such as cDNA library preparation, microarrays, RT-PCR and other PCR-based assays. It is also critical for high-throughput transcriptome analysis and high-throughput sequencing. A rapid and efficient isolation method to obtain high purity sample with maximal yield of non-degraded RNA is the key to the success of all these analyses.
Due to the delicate nature of RNA, the RNA purification process consists of a variety of unique challenges, one of which is ribonuclease (RNAse) contamination. RNAses are abundant in the environment and even a trace amount of RNase contamination can sabotage RNA-based experiments. Several precautions such as the use of RNase-free reagents, dedicated pipettes, glassware, gloves, and working in an RNAse-free environment need to be followed to achieve a good RNA yield.
Often minute amounts (low viral load per ml, typically 7812062235 particles/ml) of viral RNA need careful extraction from samples such as tissues, nasopharyngeal/oropharyngeal swabs, sputum, blood, plasma, or other body fluids. Viral RNA might also be extracted from water or other environmental samples. Sometimes investigators might need to quantify the viral particles contaminating medicinal products such as vaccines. Since the viral load of these biological, medicinal, and environmental samples is usually exceptionally low, RNA isolation consists of two major steps - virus concentration followed by RNA extraction. Viral concentration is usually achieved by applying various precipitation, flocculation, and filtration techniques.
The three most common RNA extraction strategies are:
- Organic extraction method
- Spin-column based method
- Magnetic bead-based method
The Organic Extraction Method
The organic extraction method is the most tried-and-tested method for RNA extraction and removal of cellular proteins. Here RNA isolation is achieved through organic extraction followed by RNA precipitation. This technique involves lysis/extraction in a monophasic solution of phenol and guanidine isothiocyanate. Chloroform is then added. The phenol-chloroform mixture is immiscible with water. Therefore, when centrifuged, the sample forms two distinct phases. The lower (organic) phase and phase interface contain denatured proteins, while the less-dense upper (aqueous) phase contains the RNA. The aqueous phase containing the RNA is carefully removed by pipetting (with care not to touch the interface or organic phase, as this can contaminate the sample). The RNA is then precipitated with isopropanol and rehydrated for further analysis.
Organic extraction protocols are well-established and are useful for most sample types. Proteins are rapidly denatured, and RNA is quickly stabilized. The process is scalable and can be completed in 30-60 minutes. However, this method is not amenable to high-throughput processing and is difficult to automate. New users find the phase separation and careful pipetting of aqueous phase challenging to master. Chemical fume hood needs to be used due to the hazardous chemicals. This method also requires consideration of appropriate disposal of these chemicals. Manual handling of large number of samples is cumbersome.
The Spin-Column Based Method
The easiest and safest method, readily available in a kit format, is the spin-column based method. The binding element in spin-column systems is usually composed of glass particles or powder, silica matrices, diatomaceous earth, and ion exchange carriers. In this method, nucleic acid binding is optimized with specific buffer solutions and extremely precise pH and salt concentrations. Sample lysates are passed through the silica membrane using centrifugal force, with the RNA binding to the silica gel at the appropriate pH. The membrane containing residual proteins and salt is then washed to remove impurities, and flow-through is discarded. RNA is subsequently eluted with RNase-free water.
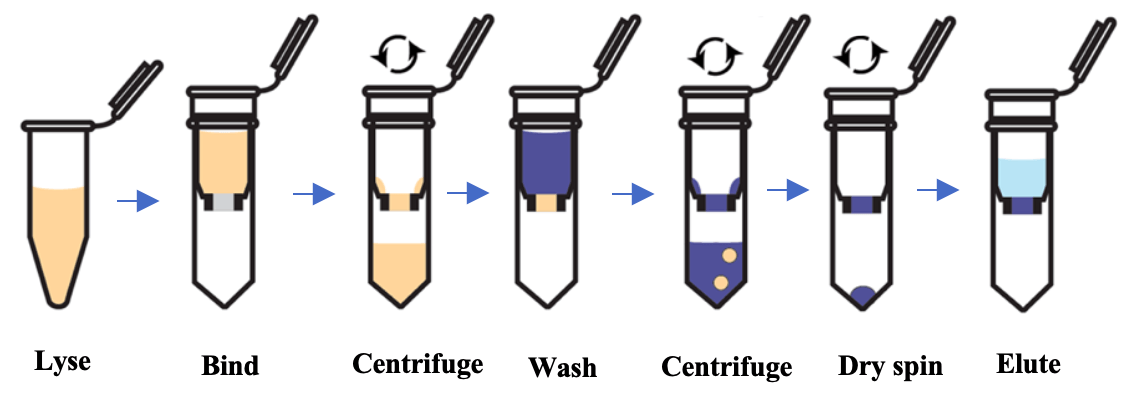
Fig 1: Schematic of RNA isolation using spin-column technology
Column-based RNA extraction is one of the best techniques among the options available, playing a vital role in ion exchange methods, as it provides a robust stationary phase for a rapid and reliable buffer exchange and thus nucleic acid extraction. This method is fast and reproducible, and its main drawback is the need for a small centrifuge. Vacuum-based systems can also be used in place of centrifugation to separate impurities. Researchers can also combine the organic extraction method with the spin column method for faster and greater RNA yield. This method is fast (20 minutes) and amenable to large-scale and high-throughput processing, including automated methods. Protein or DNA contamination is possible if the sample amount is large or remains incompletely homogenized or lysed. Incomplete lysis can also lead to low yields of viral RNA. Automation can be complex and expensive due to need for setting up centrifugation or vacuum-based separation systems.
The Magnetic Bead Based Method
The magnetic bead based method relies on the use of magnetic beads and reagents optimized for RNA extraction. The beads have a paramagnetic core, usually coated with silica for nucleic acid binding. Sample is lysed in a buffer with RNase inhibitors and then incubated with the magnetic beads, allowing the particles to bind RNA molecules. The magnetic beads can then be quickly collected by being placed in proximity to an external magnetic field. The supernatant is removed, and beads are subsequently washed in a suitable wash buffer with removal of the magnetic field. This process can be easily repeated for multiple washes. The RNA is eluted from the magnetic beads with RNase-free water into solution, and the supernatant (containing the pure RNA) can then be transferred.
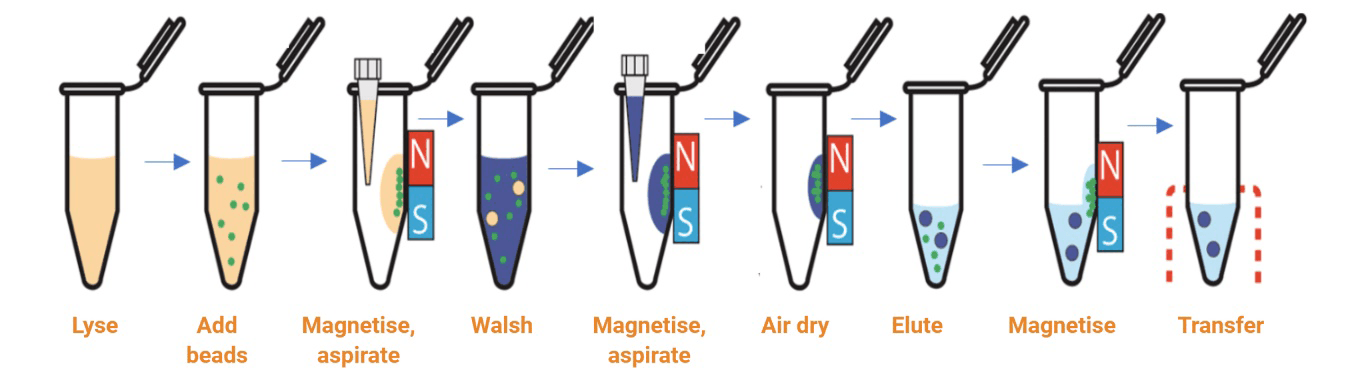
Fig 2: Schematic of RNA isolation using magnetic bead-based technology
The magnetic bead collection steps are simple and quick to perform. There is a reduced risk of clogging as no column is involved. This technique is the most amenable to scale-up, high-throughput separation, and automation. The clean-up is more effective due to the movement of the beads. However, viscous samples could impede the movement of the beads and occasionally the final sample maybe contaminated with magnetic beads. A magnetic stand is required for manual separation and a magnetic particle handler for the automation of this process.
Table 1: Summary of key differences between spin column based and magnetic bead based viral RNA isolation
| Organic extraction | Spin column based | Magnetic bead based | |
|---|---|---|---|
| Purity | Medium | High | Highest |
| Organic solvent hazardous waste | Yes | None | None |
| Difficulties | Phase separation difficult for new users | Handling several samples is tedious - Centrifuge/ vacuum required | Magnetic stand required |
| High-throughput friendly | No | Yes | Yes |
| Concern for clogging | No | Yes | No |
References
- Ali N, Rampazzo RCP, Costa ADT, Krieger MA. Current Nucleic Acid Extraction Methods and Their Implications to Point-of-Care Diagnostics. Biomed Res Int. 2017; 2017:9306564.
- Burgener M, Candrian U, Gilgen M. Comparative evaluation of four large-volume RNA extraction kits in the isolation of viral RNA from water samples. Journal of Virological methods, 2003, 108(2): 165-170.
- Kok T, Wati S et al, Comparison of six nucleic acid extraction methods for detection of viral DNA or RNA sequences in four different non-serum specimen types. J Clin Virol. 2000 Feb;16(1):59-63.
- Hjelmsø MH, Hellmér M, Fernandez-Cassi X, Timoneda N, Lukjancenko O, Seidel M, et al. (2017) Evaluation of Methods for the Concentration and Extraction of Viruses from Sewage in the Context of Metagenomic Sequencing. PLoS ONE 12(1): e0170199.
- Seah C, Chow V, Chan Y, Doraisingham S (1995) A comparative, prospective study of serological, virus isolation and PCR amplification techniques for the laboratory diagnosis of dengue infection. Serodiagnosis and Immunotherapy in Infectious Disease 7: 55-58.
- Comparison of five viral nucleic acid extraction kits for the efficient extraction of viral DNA and RNA from cell-free samples. DOI: 10.15761/TiM.7812062235.