Utilizing Nucleotide-Based Methods for Optimal Detection
Utilizing Nucleotide-Based Methods for Optimal Detection
The tactical application of oligonucleotide and aptamer assays in drug discovery
Principles for Oligo and Aptamer Methods
Aptamers are single stranded DNA or RNA oligonucleotides that have high affinity and specificity towards a wide range of target molecules. Since aptamers exist in nature but can also be artificially isolated from pools of random nucleic acids through an in vitro selection process - Selective Evolution of Ligands by Exponential Enrichment (SELEX), they can potentially bind a diverse array of compounds. They have therefore attracted immense interest and found wide applications in a range of areas.
They also offer significant advantages as recognition elements over traditionally used antibodies. They are small in size, chemically stable and cost effective. Aptamers bind to their targets with high affinity, particularly with macromolecules (e.g., proteins), with dissociation constants in the picomolar to nanomolar range. More importantly, aptamers offer remarkable flexibility and convenience in the design of their structures. Their conformational adaptability provides better affinity and selectivity than antibodies. Since aptamers can be generated by a chemical process, thereby bypassing the use of biological systems in their synthesis, aptamers against non-immunogenic agents or compounds that may be toxic to cells can also be developed. While antibodies generally have multiple binding sites, most aptamers have limited specific binding sites. Thus, there is potentially far less nonspecific binding of non-target compounds when aptamers are utilized in immunoassays as antibody alternatives. This necessitates a minimum blocking strategy and provides a more reliable detection signal for the presence of the target compound. Most importantly, unlike antibodies, aptamers can be generated against virtually any kind of biomolecules, including lipids, sugars and even small molecules.
Analogous to immunoassays based on the antigen-antibody interaction, aptamer-based bioassays can adopt different assay configurations to transduce bio-recognition events. Accordingly, various assay configurations have been designed and reported. Nevertheless, the majority of these designs fall into two categories (Fig. 1.): a. Single-site binding and b. Dual-site binding. For small molecular targets, nuclear magnetic resonance studies have indicated that they are often buried within the binding pockets of aptamer structures (Fig. 1A), leaving little room for the interaction with a second molecule. Hence, small-molecule targets are often assayed using the single-site binding configuration.
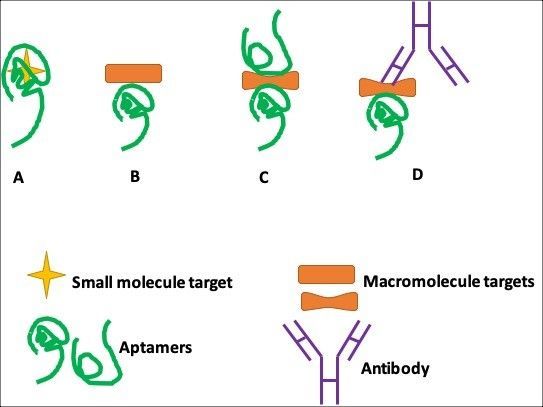
Fig. 1. Aptamer-based assay formats. (A) Small-molecule target buried within the binding pockets of aptamer structures; (B) single-site format; (C) dual-site (sandwich) binding format with two aptamers; and, (D) ‘‘sandwich’’ binding format with an aptamer and an antibody.
In contrast, protein targets are structurally complicated, allowing the interplay of various discriminatory contacts (e.g., stacking, shape, complementarity, electrostatic interactions, and hydrogen bonding). As a result, protein targets can be assayed via both single-site binding (Fig.1B) and dual-site binding (Fig.1C and 1D). The dual-site binding (aptamer only) assay relies on the availability of a pair of aptamers that bind to different regions of the same protein.
A very important immunoassay that has found varied applications in research, food safety analysis, and diagnostic medicine is the Enzyme-linked immunosorbent assay (ELISA). One approach of ELISA, commonly referred to as sandwich ELISA, involves the simultaneous use of two antibodies or analyte-binding receptor proteins to capture the analyte of interest and to report target detection. Following this idea of sandwich ELISAs, the first dual site-binding, mixed ELISA–aptamer-based bioassay was described by Drolet and colleagues in 1996 (Fig. 2.). This new detection method was appropriately coined ELONA (Enzyme-Linked Oligonucleotide Assay) in which the reporting antibody/ protein of ELISA is substituted for a fluorescein-tagged aptamer specific for detecting the target of interest (Fig. 1). Drolet et al. used a fluorescein-tagged nuclease-resistant aptamer derived through the SELEX process against human VEGF (hVEGF) and a monoclonal hVEGF capture antibody as a model. Signal was collected for quantitation of hVEGF in human sera using a chemiluminescent alkaline phosphatase detection system after final incubation with alkaline-phosphatase-conjugated anti-fluorescein Fab fragments. The authors showed that error, precision, accuracy, interference, and specificity analyses show equivalence to a conventional sandwich ELISA assay, demonstrating that ELONA represents a viable alternative to ELISA in clinical and research applications.
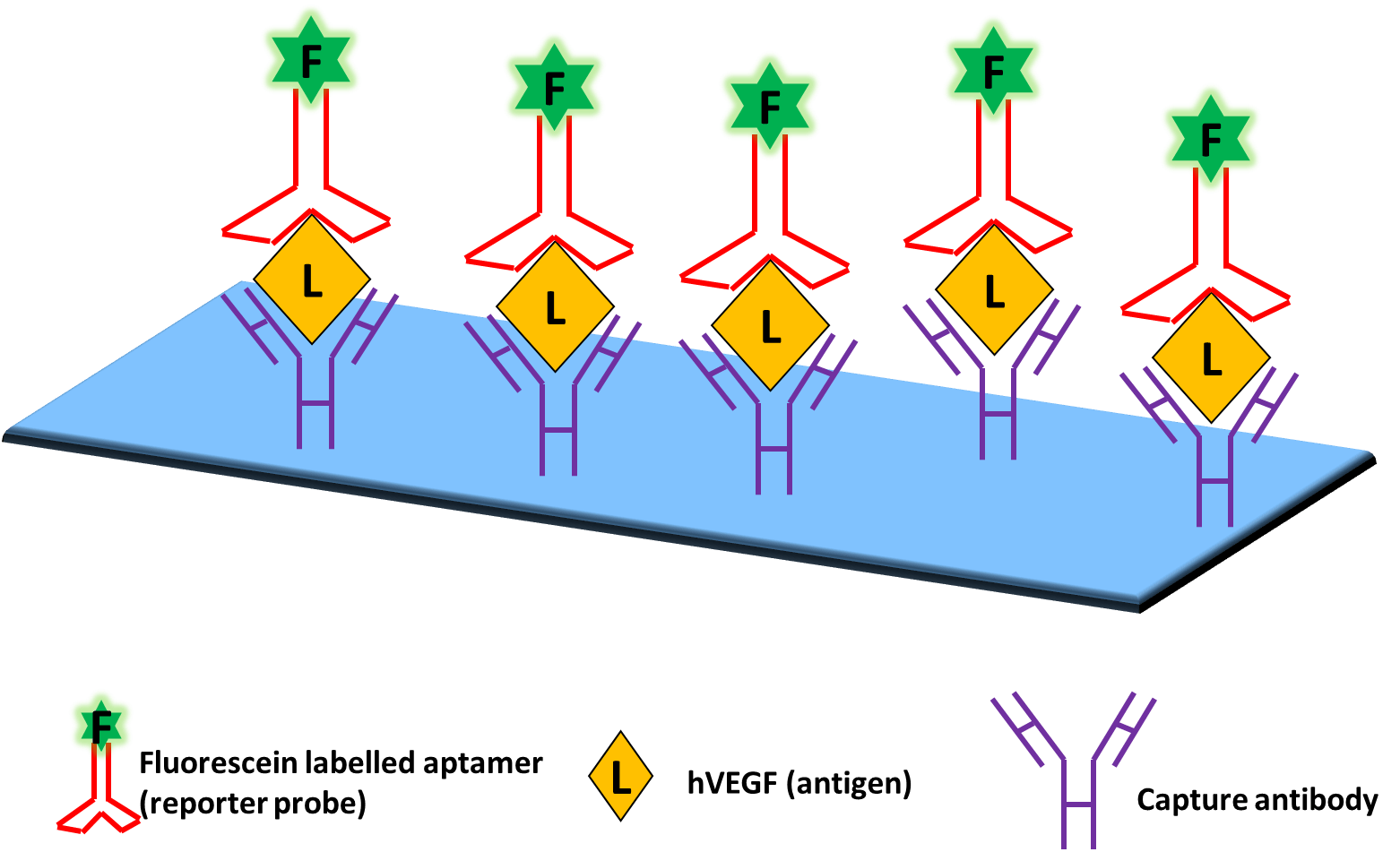
Fig. 2. First sandwich ELONA described by Drolet et. al. (1996). This assay made use of antibodies to capture hVEGF and fluorescein-labelled aptamers as a reporter
To further exclude the use of antibodies in sandwich assays, Vivekananda and Kiel devised a method that employs aptamers as the target capturing and reporting elements. This technique was coined ALISA (Aptamer-Linked Immobilized Sorbent Assay). It was applied to examine the specificity of an aptamer against an antigen associated with Francisella tularenis, a bacteria which causes tularemia or rabbit fever, an infectious endemic disease. ALISA is also referred to as Enzyme-linked aptamer sorbent assay. ALISA relies on two different aptamers generated against one target. One aptamer attached to the surface (chip, microtiter plate, nanoparticles etc.) is called the capture probe while the second aptamer labelled with the reporter is called the reporter probe. The reporter probe gives a signal upon binding to the target. Reporter probes are often conjugated with signalling moieties (e.g., fluorophores, enzymes or nanoparticles). Generally speaking, capture and reporter probes have different nucleic acid sequences; however, in limited cases, some proteins (e.g., dimeric) contain two identical binding sites, thus allowing the use of a single aptamer for the sandwich assay.
ELONA, also known as Enzyme-linked aptamer assay (ELAA), can also be used in the direct or indirect competitive bioassay configuration to detect small amounts of analyte. Similar to competitive ELISA, in competitive ELAA, the labelled and unlabelled analyte compete for a limited amount of binding aptamer. In the indirect competitive ELAA after allowing binding of the labelled analyte to the microtiter plate and blocking with an appropriate blocking agent, suitably labelled aptamer probe along with unlabelled analyte is added. After binding completes, a suitable read-out method is used to collect the signal generated from the bound labelled aptamer probe. In the direct competitive assay, the labelled aptamer is adsorbed on the microtiter plate. After suitably blocking the plate, a mix of labelled and unlabelled analyte is added and the signal is collected for quantification.
Outline of Steps and Materials in the Experimental Workflow
The ELONA method consists of the following steps:
Immobilization of the analyte/labelled aptamer on the assay platform
This step involves binding of either the analyte or capture probe, depending on the assay format being employed, to an appropriate platform such as a microtiter plate or chip. Immobilization can be achieved through covalent binding, adsorption, affinity coupling etc. After this step, is a blocking step that ensures all non-specific binding sites are blocked.
Sample preparation
This step varies depending on the specific analyte of interest. For e.g. it could include genomic DNA from a pathogen, patient sera or blood samples, protein lysate from bacteria etc. Appropriate steps need to be followed for sample extraction as the case may be.
Sensing the analyte
In case of direct, single-site binding ELAA, where the analyte is immobilized on the plate support, this step involves addition of labelled aptamer. Following binding and washing, the signal can be detected using an appropriate method depending on the label associated with the aptamer probe, i.e. fluorophores, enzymes etc. In case of direct, single-site binding ELAA, where the capture probe is immobilized on the solid support, this step involves addition and binding with labelled analyte, signal from which can then be detected after washing. For dual site-binding/sandwich ELAAs there is an additional step where the reporter probe is added following binding of analyte to the capture probe.
As described earlier, ELAA can be run as a competitive binding assay too. An example of indirect and direct competitive ELAA for quantification of residual tetracycline (TC) from antibacterial treatment of hives during the production of honey are described below (Fig. 3). In the indirect format, the microtiter plates are first coated with TC-BSA (competitor antigen). After blocking with Hammerstein bovine casein, biotinylated aptamer and unlabelled TC are added and binding allowed. After washing Streptavidin-Horseradish peroxidase (SA-HRP) is added. Finally TMB solution (HRP substrate) is added, followed by H2SO4 to stop the reaction and absorbance is measured. In the direct competitive ELAA for TC using the same biotinylated aptamer, the plate is first coated with SA, blocked and then binding of biotinylated aptamer along with unlabelled TC is carried out. After washing, TC-HRP conjugate is added and TMB solution is used for the signal readout as earlier.
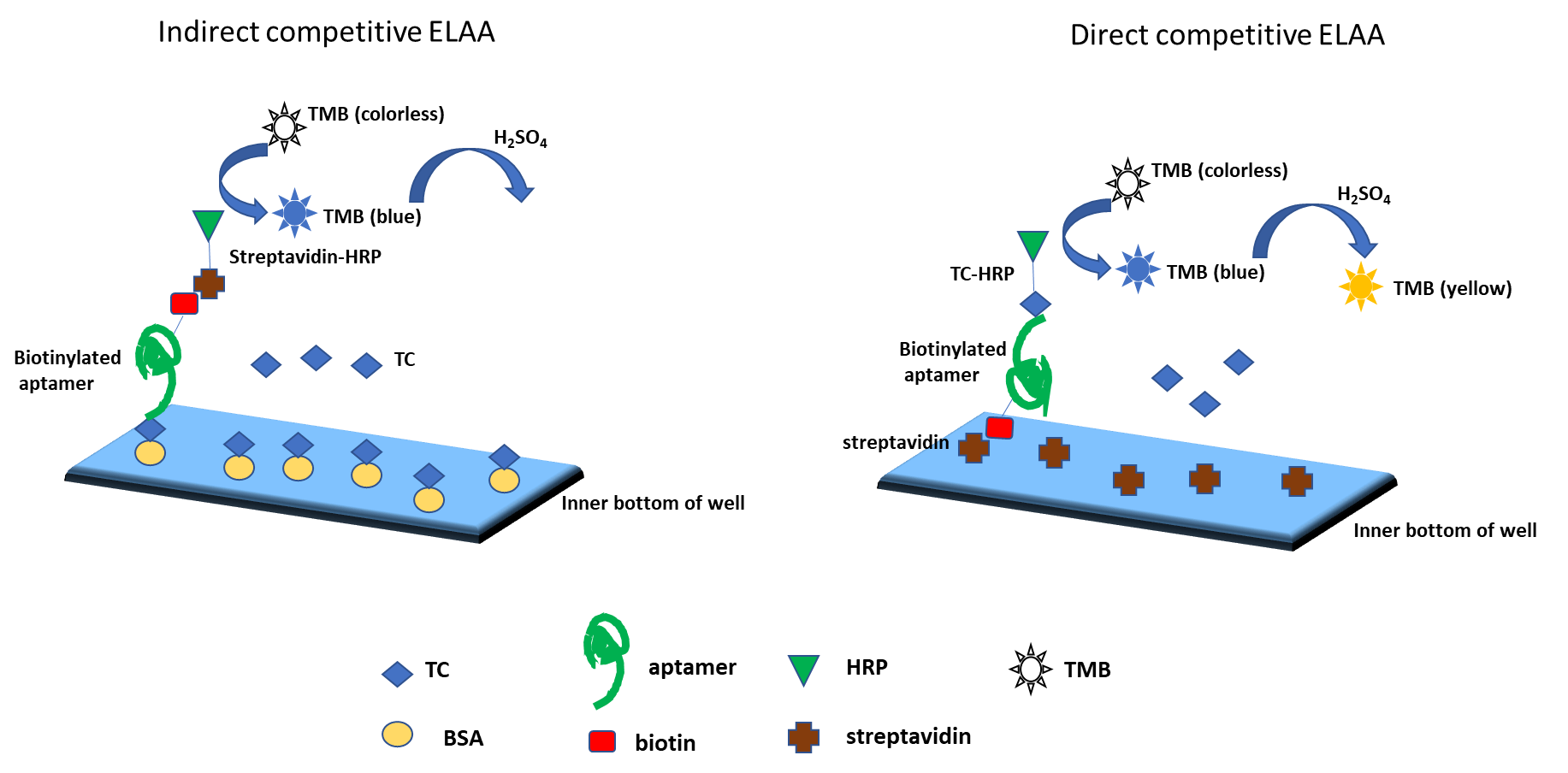
Fig. 3. Direct and indirect competitive ELAA for detecting residual tetracycline in honey
Value of This Application in the Discovery Stage of Research
Since the first reported ELAA in 1996, significant progress has been made in utilizing aptamers in detection assays.
Recently, several groups have reported aptamer-based biomarker discovery platforms with multiplexing capabilities. Gold and co-workers have described a biomarker discovery system that is capable of simultaneously measuring thousands of proteins from serum or plasma samples. Using their system, they discovered 58 potential biomarkers for chronic kidney disease. The same group, using their patented aptamers called SOMAmers, reported a large-scale study of screening serum samples to discover biomarkers for non-small cell lung cancer. These two reports showcase the significant improvements in the development of highly sensitive, aptamer-based biomarker discovery platforms.
A novel electrochemical sensor system based on two different aptamers for the detection of thrombin was developed by Ikebukuro et al. The authors integrated two aptamers recognizing different epitopes of thrombin (fibrinogen and heparin binding sites) in their sandwich assay. One of the aptamers was thiol-modified and immobilized on a gold electrode for capturing thrombin, while the other aptamer, labeled with a pyrroquinoline quinone glucose dehydrogenase, indicated complex formation.
To overcome the limitations associated with colorimetric or fluorescent readout in ELONAs, Morena et al, have combined the mid-range sample throughput potential of ELONA with the sensitivity of SYBR green-based, real-time quantitative PCR(qPCR). The use of aptamers as (RT)qPCR-amplifiable reporter molecules excludes the need for aptamer or target labelling, allowing a straightforward assay with enhanced sensitivity.
Their specific binding properties and antagonistic effect on protein function makes aptamers well suited as artificial ligands for target validation applications in context with high-throughput screening. Green et al. provided the first evidence that known small-molecule inhibitors can compete with aptamers in biochemical displacement assays. Researchers have expanded this concept to novel inhibitors of disease-relevant targets. For example, screening with inhibitory aptamers led to the identification of functionally equivalent low molecular weight compounds for the guanosine 5′-diphosphate (GDP)/guanosine 5′-triphosphate (GTP) exchange factor cytohesin-1 and the HIV rev protein. These aptamer-based competitive screening assays were successfully adopted to different readouts, such as fluorescence intensity, fluorescence resonance energy transfer, fluorescence polarization, fluorescence lifetime, radioactivity, and enzyme-linked reactions.
Along with the rapid progress of modern analytical technologies and the application of novel analytical reagents (e.g., nanomaterial-based probes), more and more aptamer-based bioassay formats are being developed. These technologies have been introduced into analytical applications, target validation, and drug discovery processes. In contrast to many other drug discovery technologies, neither target nor small-molecule labeling is required in the aptamer approach. It is especially useful when the natural ligand/substrate is unknown, too expensive to produce, or an x-ray structure is unavailable. Furthermore, these assay systems can be applied when new sites on a protein need to be explored, aiming for small-molecule drugs with potentially new mechanisms of action.
References
- Mok W, Li Y. Recent Progress in Nucleic Acid Aptamer-Based Biosensors and Bioassays. Sensors (Basel). 2008;8(11):7050-7084.
- Ikebukuro K, Kiyohara C, Sode K (2005) Novel electrochemical sensor system for protein using the aptamers in sandwich manner. Biosens Bioelectron 20:2168–2172.
- Moreno M, et al. Combined ELONA-(RT)qPCR Approach for Characterizing DNA and RNA Aptamers Selected against PCBP-2. Molecules. 2019;24(7):1213.
- Gold L , Ayers D, Bertin J, Bock C, Bock A, et al. (2010) Aptamer-Based Multiplexed Proteomic Technology for Biomarker Discovery. PLoS ONE 5: e15004.
- Drolet, D.W.; Moon-McDermott, L.; Romig, T.S. An enzyme-linked oligonucleotide assay. Nat. Biotechnol. 1996, 14, 1021-1025.
- Dong Y and Wang S, Enzyme-linked aptamer assay. Aptamers for Analytical Applications: Affinity Acquisition and Method Design, 2018 Wiley-VCH Verlag GmbH & Co. KGaA
- Somasunderam A, Gorenstein DG, Aptamers as Novel Reagents for Biomarker Discovery Applications. Translational Medic 2011. S1:001. doi:10.4172/2161- 1025.S1-001
- Proske, D., Blank, M., Buhmann, R. et al. Aptamers—basic research, drug development, and clinical applications. Appl Microbiol Biotechnol 69, 367–374 (2005).