Rethink Modifications
Rethink Modifications
ssDNA, siRNA, gRNA, LNA, BNA; the list goes on.
Overview
Advancements in biochemistry continue to add new and innovative tools to help build DNA and RNA sequences. As tools are added, it becomes increasingly difficult to determine the right tool for the right objective.
In this case; "tools” refer to modifications, or 'mods'. In Nucleotide synthesis; modifications are specialty synthetic reagents or compounds that can be introduced into a nucleotide sequence. Applying these modifications in design allows researchers to customize and manipulate various aspects of their sequence including its biochemical properties. This can help achieve a variety of goals, including improved stability, specific directional form, higher affinity, and other goals stemming from the primary application objectives of researchers.. The recent boom in oligonucleotide applications has resulted in more modifications and variations from which to choose, and more companies offering their own variations of all types of modifications. Naturally, this has begun to generate confusion in the marketplace.
Finding the right modifications to build a DNA or RNA sequence can be overwhelming, a problem that has resulted in a growing trend of laboratories dedicating valuable time and money to unnecessary and expensive modifications, sequence designs that overlook biochemistry fundamentals, and widespread misinformation. With a growing need for biochemists in nucleotide synthesis labs, there is an equal need to re-establish an understanding of the different types of modifications and the roles they play in synthesis.
- Introducing nucleotide synthesis modifications using an alternative approach
- Outlining sequence versus base modification methods
- Categorized explanations of modification types and their value
Introduction
In many cases, modification errors occur with novel sequence designs that overlook one or more biochemical principles. In other cases, errors can usually be attributed to confusion about or misunderstanding of modification chemistry in general. Before taking into account their biochemical properties, it is important to consider nucleotide modifications within the following framework;
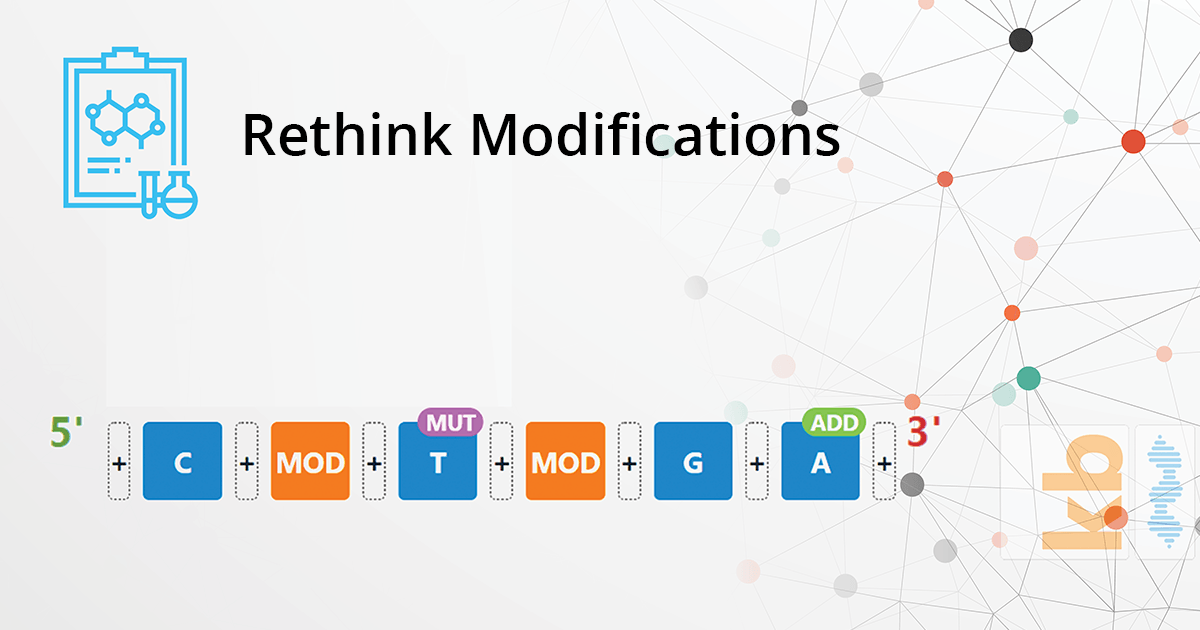
A modification can be one of the following primary types:
Sequence Modification
Base Modification
Categorizing your modification of interest as either a sequence or base mod will help establish its hierarchy of influence within your sequence design. This will also provide an organized structure to calculate its effects on the chemistry as it relates to the nucleotide synthesis project.
Sequence Modifications
A sequence modification is any mod that adds or edits features to the nucleic acid sequence. The primary distinction with this class of modifiers is that they target the structure and function of the final molecule. The demand for sequence modifications is relative to experimental challenges. For example:
Improving stability or protecting from degradation
Introducing markers for detection in analysis experiments and higher affinity
Troubleshooting nucleic acid conformation issues and directional form
Sequence Modification Subcategories
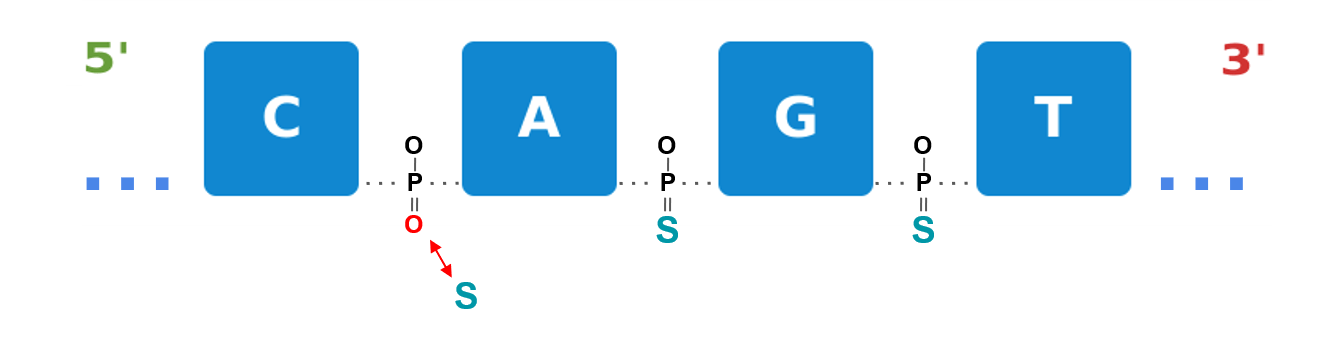
a) Backbone | Sequence Modification to the phosphate backbone structure
Example: Phosphothioation
Figure 1a: Phosphothioation substitutes the non-bridging oxygens of the phosphate backbone with sulfur atoms. This modification reinforces the molecules’ stability and resistance toward nuclease degradation
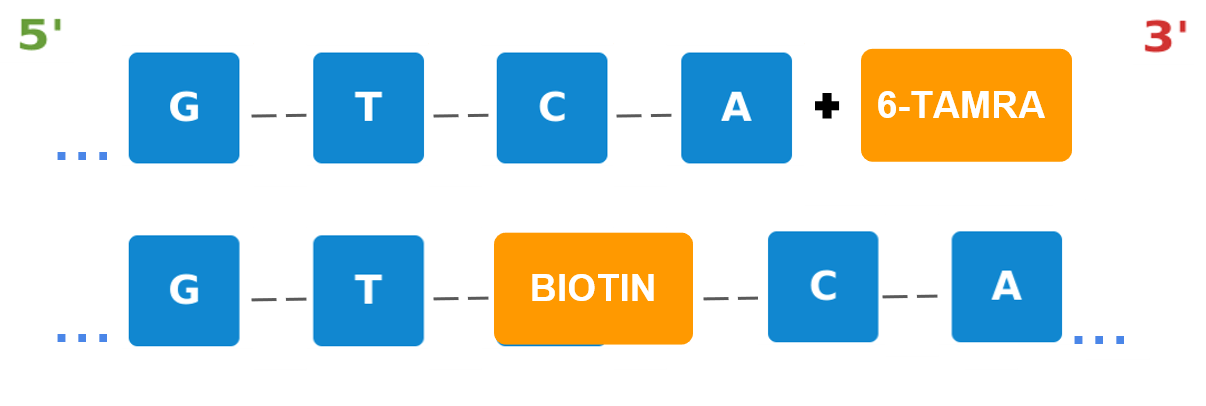
b) Chain | Sequence Modification to the chain of nucleotides via attachment of different compounds and functional groups
Example: Fluorescent labels and peptide conjugates
Figure 1b: Attaching a 6-TAMRA label to the 3’ end (top) offers a fluorescence feature to the bioreagent. Incorporating an affinity tag such as an internal-BIOTIN (bottom) can help support purification along with a variety of additional functions.
Base Modifications
Where-as sequence modifications impact the nucleic acid sequence, a base modification focuses on the amino acid-base of one or more nucleotides within the nucleic acid sequence. It can be introduced as:
- A deletion or removal of one or more nucleotides
- An insertion or addition of one or more nucleotides
- A mutation or change in the amino acid of one or more nucleotides
Base mods often address the more pivotal objectives of an application and can range from a single base mutation to a series of deletions and insertions. However simple they may seem, base mods are also routine inducers of experimental complications that require the use of complementary sequence modifications. This is where expertise in biochemistry and synthesis technology becomes crucial.
Base modifications come with a high level of uncertainty. This is especially true when working with different applications. It can be very difficult to accurately predict certain performance outcomes after base modifications are applied. We refer to these unpredictable outcomes as 'blind spots'. Conducting analysis experiments is the current standard for calculating blind spots and the integration of these analysis models into sequence design platforms is a growing area of demand.
Understanding the Available Options
Over the years, many companies have developed extensive catalogs with overwhelming numbers of modifications from which to choose. In order to effectively apply any modification - it is essential to first understand what class or function it fits into. We refer to these as the mod’s subcategory. kbDNA applied the less-is-more approach and focused on optimizing each modification in relation to its subcategory. The resulting selection consists of modifications focused on the following options:
- Affinity Tags
- Fluorescent Labels
- Structural Modifiers
- Stabilizing Assistants
- Degenerate Carrying Bases
- Peptide Conjugates
- Terminal Phosphates
We plan on following up with more detailed profiles for each modification option including their individual structures and biochemical properties. As part of our conservative approach to developing a more precise tool for building nucleotide sequences; more effort is being channeled into a valuable release of this material rather than a generic presentation. Our modification abilities and options are currently well represented in kbDNA’s nucleotide synthesis builder. The builder resource was also used to produce the figures depicted in this note and is available to everyone. We encourage researchers to explore the building resource here to learn more about the right options for their laboratories.