Cloning of Single-Chain Fv Fragments (scFv's)
Cloning of Single-Chain Fv Fragments (scFv's)
Single-chain fragment variables (scFv’s) with a molecular weight of ~25 kDa represent the smallest unit of a functional antibody with complete antigen-binding activities1 as shown in Figure 1. scFv’s (single-chain fragment variables) are generated by fusion of immunoglobin heavy chain (VH) and light chain (VL) variable domains connected by flexible polypeptide linker2. scFv’s are easily expressed in functional form for improvement of properties such as increased affinity, solubility, and modulation of specificity in various expression models such as E. coli, yeast, and mammalian cells. Among all the available expression systems, the bacterial expression system is most commonly utilized for the production of scFv antibody fragments. This protocol provides a quick guide related to the main aspects of scFv cloning.
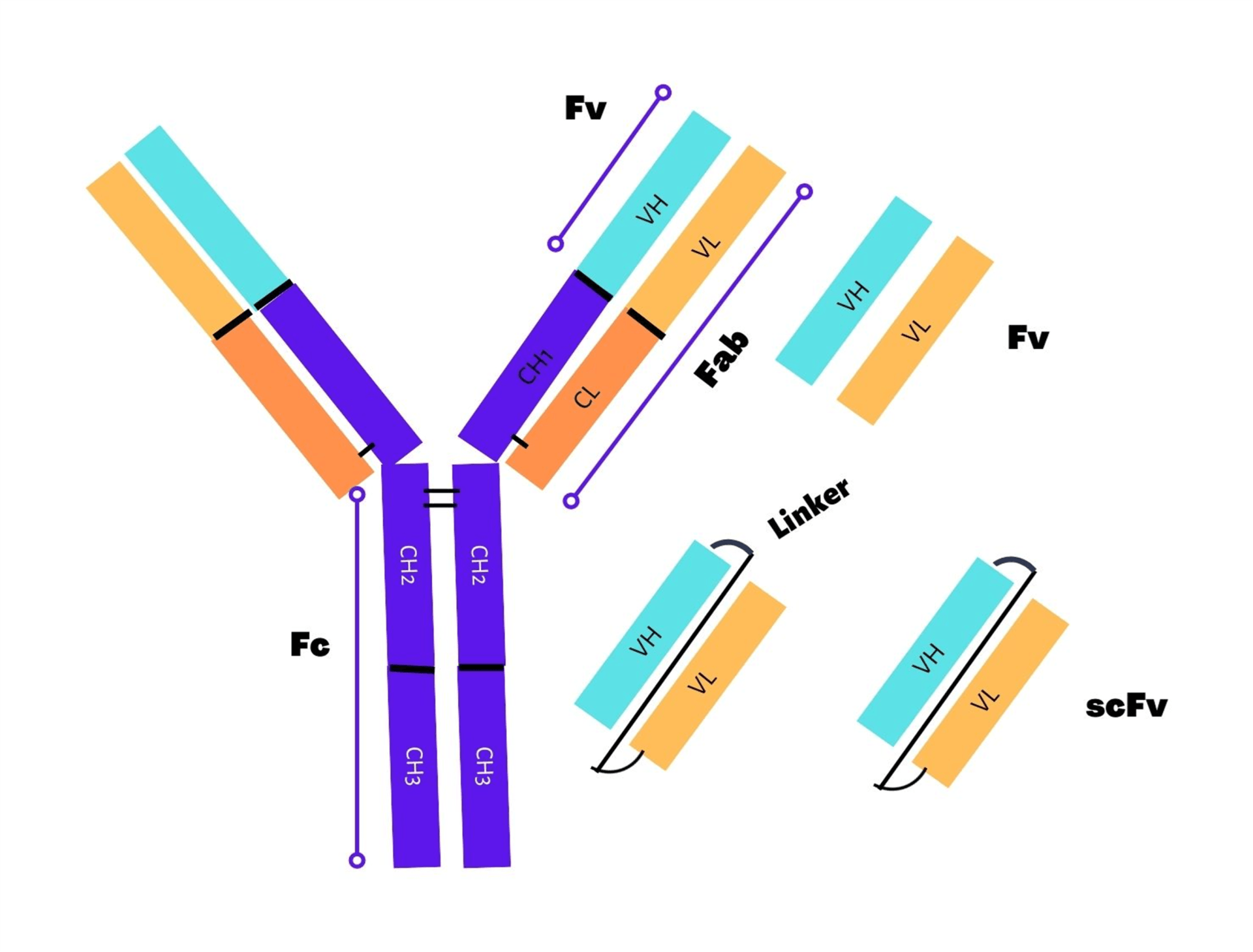
Orientation of variable domain and location of tag in scFv fragment:
During construction of the scFv (single-chain fragment variable), there is no preferential orientation of one domain over the other variable domain. The orientation can be either VH-linker-VL or VL-linker-VH and both orientation types are equivalent. Owing to its nature as independent folding entities, ScFv fragments can be fused indistinctively on either end with other protein domains or epitope tags. For example, the N-terminal end of scFv’s has been tagged extensively with FLAG tag3, while the C-terminal end has been frequently tagged with PHEN1 in many vectors4. Furthermore, purification tags such as HIS tag, Streptag etc. must be located on the C-terminal side to prevent purification of truncated proteins. In order to make scFv more accessible, purification tags are often located first, followed by C-terminal epitope tags. In the case of phage display, free scFv are allowed to secrete in the periplasmic space using a combination of epitope tag plus amber codon between the scFv and the phage coat protein for binding and analysis purpose. In a recent study, P17 tag (a 33-residue peptide) was utilized as tag for improvement of scFv’s solubility (11.6-fold increase) and thermal stability5.
Length of linker and its sequence:
A flexible short peptide or linker must be designed once the orientation is finalized along with tagging with proteins or epitope tags as it plays a vital role in stabilizing the antibody. Earlier studies reported 3.5 nm (35 Ǻ) length for peptide linker between the C-terminal and N-terminal end of scFv’s variable domains without affecting the folding ability6. The length of the linker is critical to ensure the correct folding of different domains in scFv’s. If the length of the linker is too short (5-10 amino acids long), the physical association between the variable domains is hindered and leads to the formation of multimers (diabodies, tribodies, etc.)7. On the other hand, linkers that are too long would favor weak domain association or susceptibility to proteolysis. Therefore, choosing optimum length linkers is pertinent while constructing scFv’s or biospecific antibodies. In general, a linker length of>12 amino acid residues allows sufficient distance between the variable domains for interactions and monomer formation. Currently, linkers of length 15-20 amino acids long are being utilized to promote physical interactions of variable domains and prevent formation of dimeric forms.
Apart from the linker length, in order to design a viable and flexible peptide linker, their amino acid composition is also considered crucial. For example, hydrophilic amino acid in the linker sequence is essential to avoid intercalation of peptide within or between the variable domains throughout the protein folding8. Currently, amino acid motif (G4S) n (G: glycine, S: serine; G4S: four glycine and one serine residue) is most extensively utilized for linkers designs owing to its role in granting conformational flexibility, solubility improvement and minimal immunogenicity9. Most commonly utilized are multimers of pentapeptide (G4S) such as 15-mer (G4S)310, 18-mer GGSSRSSSSGGGGSGGGG11, and 20-mer (G4S)412. Apart from the Gly-Ser linker, other charged amino acid residues such as lysine and glutamic acid, sequences with functionalities (sequence containing a Cre-Lox recombination site) to enhance the scFv properties such as solubility.
Cloning methodology:
The most common method for the construction of scFv’s using either of the orientations include PCR assembly. In this method, VH and VL genes are inserted directly into the plasmid or phagemid through in vitro recombination after the PCR. This method allows cloning of variable domains of the antibody without prior information regarding amino acid composition and nucleic acids. Alternatively, sequential cloning or combinatorial infection are other methods being utilized for scFv (single-chain fragment variable) construction. A two-step overlapping PCR also known as Splicing by Overlap Extension (SOE-PCR) has been usually employed for cloning of the scFv. Herein, VH and VL domains are assembled using a single step of PCR assembly after amplification and gel-purification (Figure 2). Thereafter, using three-fragment assembly PCR (addition of linker primer) or by overlap of the two inner primers, linker is generated. An equal amount of VH and VL domains (50-100 ng of each domain) are utilized in the PCR assembly method in order to avoid preferential amplification of any one domain. There are two types of primers (outer and inner) being utilized in assembly PCR. The outer primers include the primers used for amplification or new pair of extension primers. The extension primers help extend the sequence from both sides of the scFv either by adding a restriction site or amplification of assembly independently for library creation. Sometimes, a few PCR cycles are performed to assemble the two variable domains before adding the outer primers. Although, the advantage of this step is not clearly established related to scFv generation.
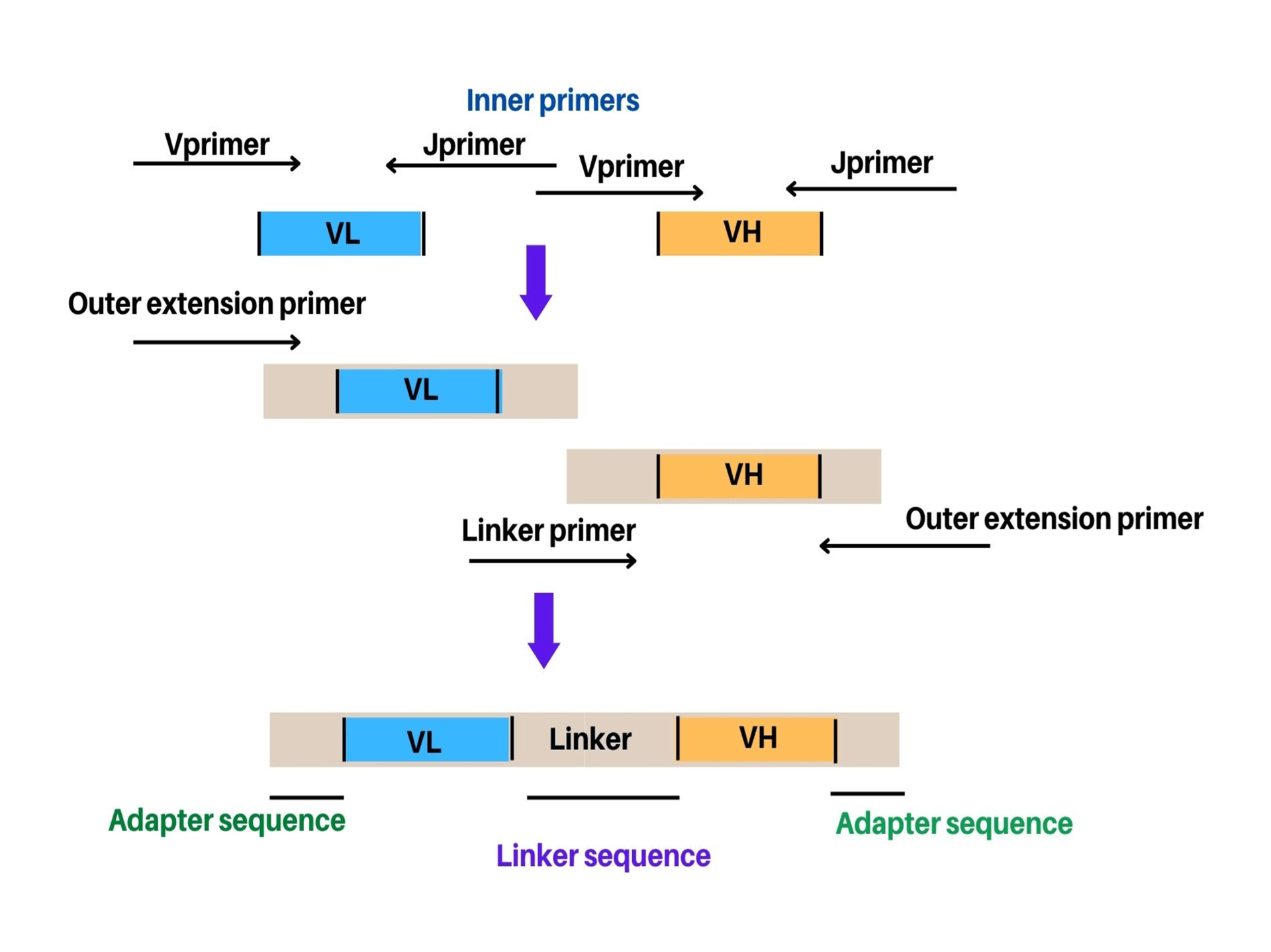
Example of assembly reactions:
The most common assembly which have been extensively utilized include the classical (G4S)3 linker either as a three-fragment assembly or a two-fragment assembly13. The classical approach for the two-fragment assembly in scFv construction is shown below:
| Translation | G G G G S G G G G S G G G G S |
|---|---|
| Linker primer | GGTGGAGGCGGTTCAGGCGGAGGTGGCTCTGGCGGTGGCGGATCG |
| Inner primer (s) | GGCGGAGGTGGCTCTGGCGGTGGCGGATCG-Vstart |
| Inner Primer (rc) | Jend-GGTGGAGGCGGTTCAGGCGGAGGTGGCTCT |
| Note: | s – sense primer, r – reverse primer, rc - reverse complement of reverse primer. |
References
- Brinkmann U., Kontermann R.E. The making of bispecific antibodies. MAbs. 2017; 9:182–212. doi: 10.1080/19420862.2016.1268307.
- Ahmad Z.A., Yeap S.K., Ali A.M., Ho W.Y., Alitheen N.B., Hamid M. scFv antibody: Principles and clinical application. Clin. Dev. Immunol. 2012; 2012:980250. doi: 10.1155/2012/980250.
- Krebber, A., Bornhauser, S., Burmester, J., Honegger, A., Willuda, J., Bosshard, H. R., & Pluckthun, A. (1997). Reliable cloning of functional antibody variable domains from hybridomas and spleen cell repertoires employing a reengineered phage display system. J Immunol Methods, 201(1), 35–55.
- Hoogenboom, H. R., Griffiths, A. D., Johnson, K. S., Chiswell, D. J., Hudson, P., & Winter, G. (1991). Multi-subunit proteins on the surface of filamentous phage: methodologies for displaying antibody (Fab) heavy and light chains. Nucleic Acids Res, 19(15), 4133–4137.
- Wang, Y., Yuan, W., Guo, S. et al. A 33-residue peptide tag increases solubility and stability of Escherichia coli produced single-chain antibody fragments. Nat Commun 13, 4614 (2022). https://doi.org/10.1038/s41467-022-32423-9.
- Huston J. S., M. Mudgett-Hunter, M. S. Tai et al., “Protein engineering of single-chain Fv analogs and fusion proteins,”Methods in Enzymology, vol. 203, pp. 46–88, 1991.
- Gil, D.; Schrum, A.G. Strategies to stabilize compact folding and minimize aggregation of antibody-based fragments. Adv. Biosci. Biotechnol. (Print) 2013, 4, 73–84.
- Argos P., “An investigation of oligopeptides linking domains in protein tertiary structures and possible candidates for general gene fusion,” Journal of Molecular Biology, vol. 211, no. 4, pp. 943–958, 1990.
- Wang Q, Chen Y, Park J, Liu X, Hu Y, Wang T, McFarland K, Betenbaugh MJ. Design and Production of Bispecific Antibodies. Antibodies (Basel). 2019 Aug 2;8(3):43. doi: 10.3390/antib8030043.
- Huston, J. S., Levinson, D., Mudgett-Hunter, M., Tai, M. S., Novotný, J., Margolies, M. N., Ridge, R. J., et al. (1988). Protein engineering of antibody binding sites: recovery of specific activity in an anti-digoxin single-chain Fv analogue produced in Escherichia coli. Proceedings of the National Academy of Sciences of the United States of America, 85(16), 5879–83.
- Andris-Widhopf, J., Steinberger, P., Fuller, R., Rader, C., & Barbas, C. F. (2011). Generation of human scFv antibody libraries: PCR amplification and assembly of light- and heavy-chain coding sequences. Cold Spring Harbor protocols, 2011(9).
- Schaefer, J. V, Honegger, A., & Pluckthun, A. (2010). Construction of scFv Fragments from Hybridoma or Spleen Cells by PCR Assembly. (R. Kontermann & S. Dübel, Eds.).
- Marks, J. D., & Bradbury, A. (2004). PCR cloning of human immunoglobulin genes. Methods in molecular biology (Clifton, N.J.), 248, 117–34.