Microarrays Revisited [2023]
Microarray Protocol
Microarray technology represents a versatile and high-throughput technology to explore genome and parallel gene expression for thousands of genes with known and unknown functions using a variety of biological samples1,2
Overview:
In microarray technology, millions of probe molecules’ microspots are immobilized on a miniaturized solid support or chip in an array format, which are then exposed to study samples (cells, tissues, and bodily fluids) containing the corresponding target molecules. The probes utilized in this microarray technique include protein, antibody, cDNA, oligonucleotides etc., while the target molecules are DNA or RNA isolated from the biological samples (cells/tissues). The complementary base pairing of chip-immobilized fragments and target molecules produces fluorescence which is detected and quantified using a specialized machine. Moreover, the microarray technique allows simultaneous rapid detection and quantification of proteins or nucleic acids within a single reaction in an efficient manner along with comparative genomics analysis. Furthermore, it also allows the detection of specific DNA sequences (for example, single nucleotide polymorphisms or SNPs) useful for genome-wide association studies. The advantage of this technology includes its elimination of the need for sequencing, and its tremendous cost reduction unlike conventional large studies. Currently, there are different microarray formats available for the detection and analysis of specific biomolecules, namely, DNA microarrays, protein (antibody) microarrays, cell microarrays and tissue microarrays. These microarray formats either alone or in combination with other microarray techniques are being utilized for biological research and clinical studies. Using these techniques, one can examine the cell profiling treated with certain stimulators, gene expression profiling in response to drugs, serum analysis for diagnostic information and assessment of protein families, to name a few. The applications of microarray-based technology further extend to biomedical research, drug discovery and development, medical diagnostics to toxicogenomics.
Despite the variations in available protocols, the five major steps which are in essential in the microarray process include:
- Synthesis of capture element (probes),
- Solid support surface or chip preparation,
- Capture elements (probes) immobilization using a robotic arrayer onto the solid support,
- Immobilized capture elements binding with the target molecule present in the biological samples, and
- Finally, target/capture element complex detection and quantification.
Deeper Dive:
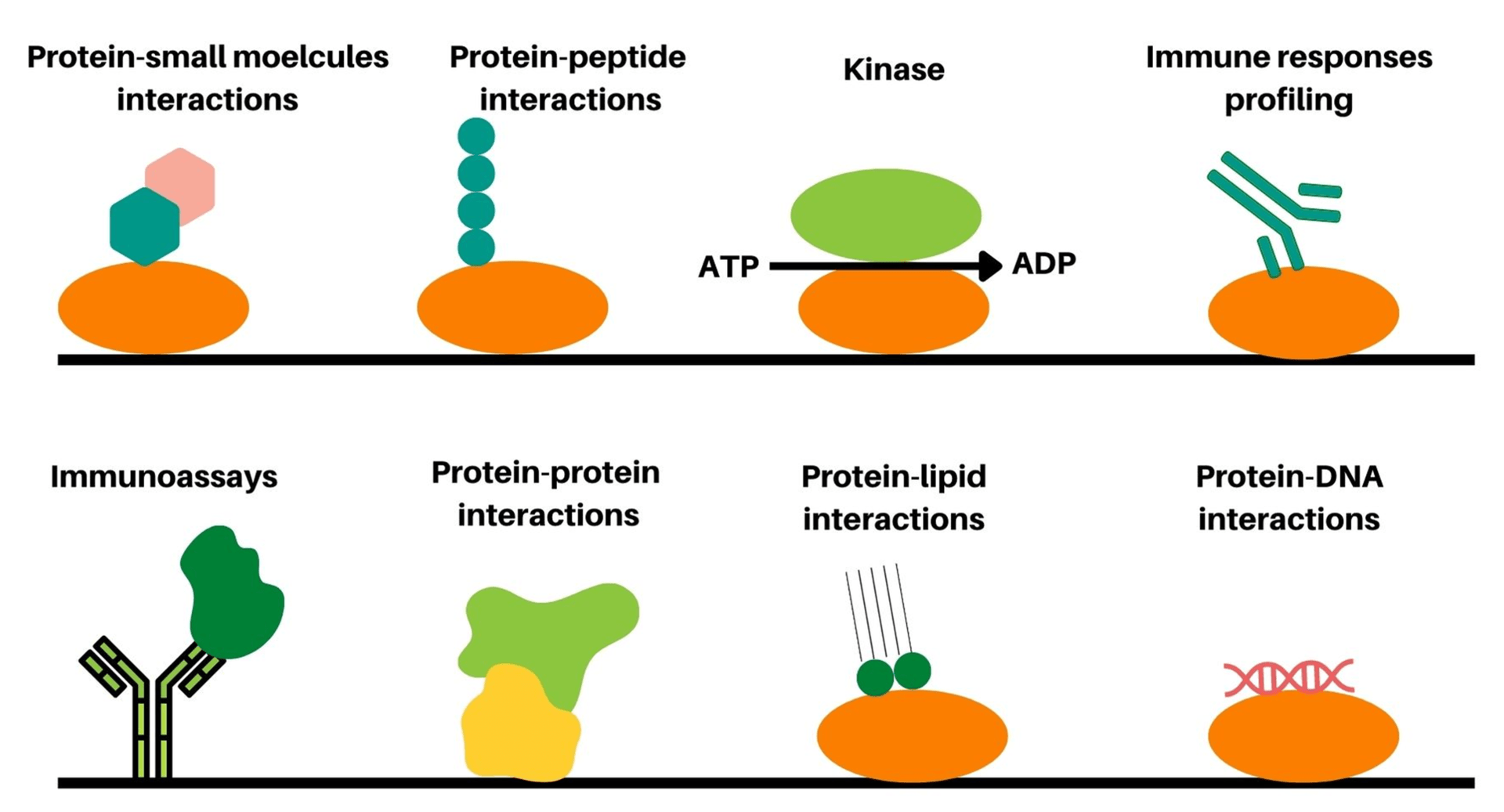
DNA microarray technology was the first developed microarray technique for the detection of genes and genome analysis, however; little information related to proteins and their functions are provided using this technique. In this context, protein microarray, or protein chip was further developed as high throughput technique to detect different proteins, protein functions, proteins interactions, protein interactions with small molecules, proteins kinase substrates, and proteomic analysis in a single experiment3.4[Figure 1]. Currently, in order to study the biochemical activities and functions of proteins, two protein microarray formats such as analytical and functional protein microarrays are mainly utilized. Analytical protein microarrays or antibody microarrays represents the most powerful multiplexed high throughput detection technologies for identifying specific proteins with high specificity and affinity. On the other hand, functional protein microarrays employ protein domains or full-length functional proteins for the identification of protein interactions with other biomolecules, immune responses, and biochemical activities. Nevertheless, both classes of protein microarrays have shown tremendous progress in the last decade in terms of sensitivity, specificity, and expanded applications.
The antibody microarray is the most representative type of protein microarray wherein antibodies are employed as the capture elements and arrayed on glass surfaces with high density for further specific detection of proteins of interest with high specificity. Herein, immobilized antibodies are tagged with fluorescent dyes namely, Texas Red, Cy3, Cy5, FITC or biotin-horseradish peroxidase enzyme systems for specific detection and visualization. In antibody microarray techniques, we employ the same well-established principles such as ELISA based immunoassays or Western blot-based immunoassays for detection of proteins, however; various advantages are offered by these microarray techniques. The advantages of antibody microarray techniques include one step detection of multiple proteins, increased number of data points for better detection accuracy, and reduced sample and antibody volume requirements. Additionally, ability to rapidly spot limitless copies using a robotic arrayer once the microarray is designed and all components are validated presents another major advantage for working with large sample groups.
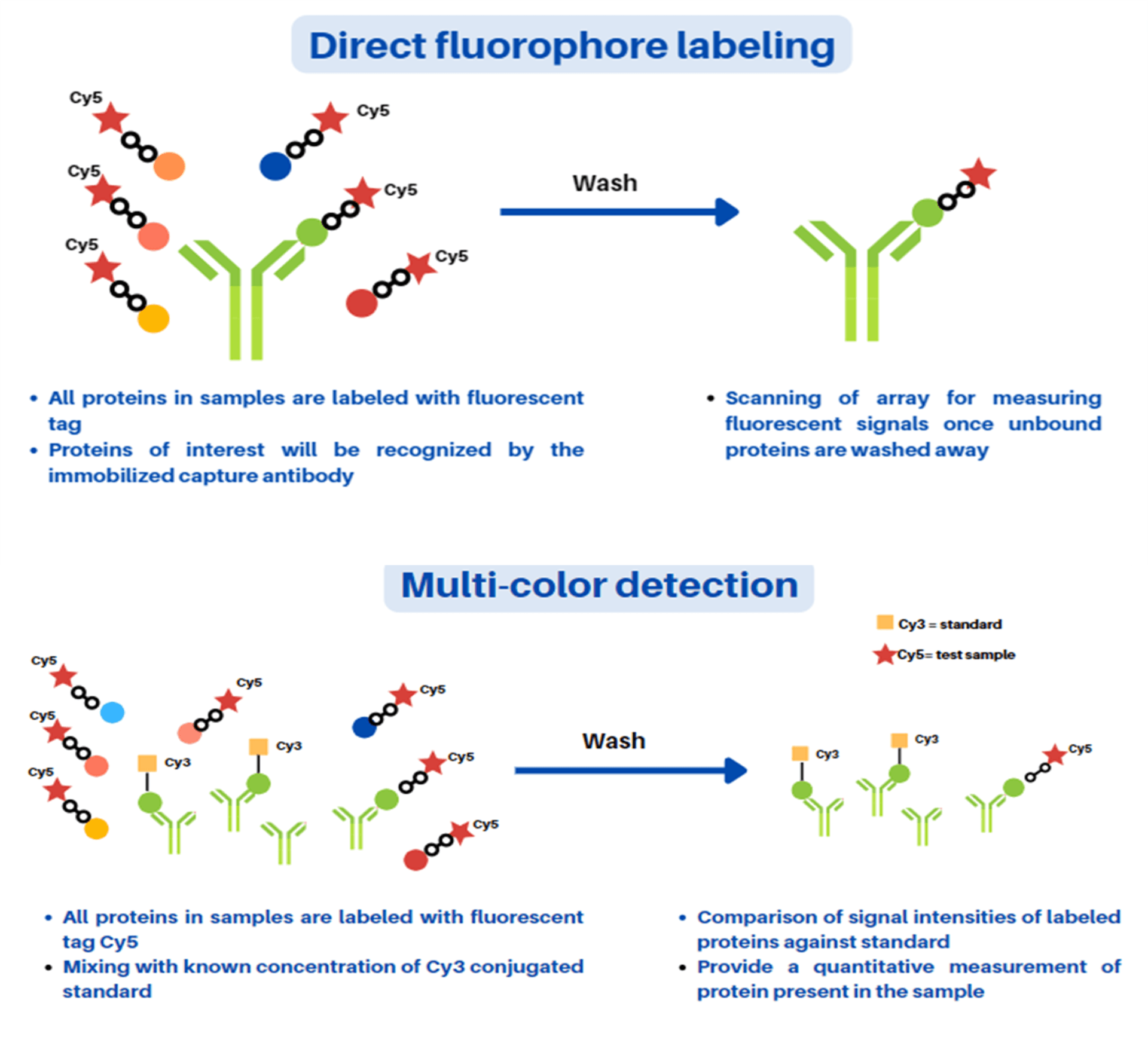
In this protocol, we have highlighted mainly two formats of the antibody microarray, namely direct labeling and fluorescence-linked immunosorbent assay (FLISA). In both these formats, specific antibodies are employed as the capture element. However, in case of direct labeling microarray technique, all proteins present in the sample mixture are labeled prior to its capture unlike in the FLISA technique. In the FLISA microarray technique, the untreated proteins present in the sample mixture are captured first and followed by their detection using a biotinylated primary antibody and fluorophore-labeled secondary antibody. In both types of microarray techniques, the employed antibodies should be highly purified with minimal cross-reactivity, and able to work well in traditional ELISA formats. Firstly, a predetermined optimal concentration (usually 300-500 µg/ml) of antibody solutions are prepared followed by the preparation of spotted solid support or chip through assembly into a print plate or microtiter plate using a robotic arrayer. Generally, up to 500 different antibodies per array can be spotted and immobilized for comparison as per the researcher’s requirement using six to ten replicates of antibody. After the spotting and immobilization of antibodies, blocking agent bovine serum albumin (BSA) solution treatment is given for at least 1 hour to block the non-specific reactions. Thereafter, the biological samples of interest such as urine, serum, cell lysate etc. are exposed to the immobilized miniaturized solid support. Direct labeling antibody microarray format-based detection is depicted in Figure 2 (A and B). In this method, all proteins present in a sample mixture are labeled with either biotin or fluorophore prior to its incubation with the capture element. Furthermore, quantitative measurement for determination of relative amounts or expression levels measurement of detected proteins are performed after washing of arrays followed by scanning for data analysis. In an alternative strategy, quantitative measurement of exact expression levels by comparing different signal intensities of sample molecules can be carried out using established calibrated protein standards labeled with distinct fluorophores. The direct labeling antibody microarray technique is simple to use and requires only one specific antibody per target. However, increased background signal due to labeling of proteins in the sample mixture (for e.g., albumin in serum) represents a major challenge in detection using this technique. The concentration of the background protein (approximately 100 ng/ml) with respect to target protein concertation in serum reduces the detection sensitivity
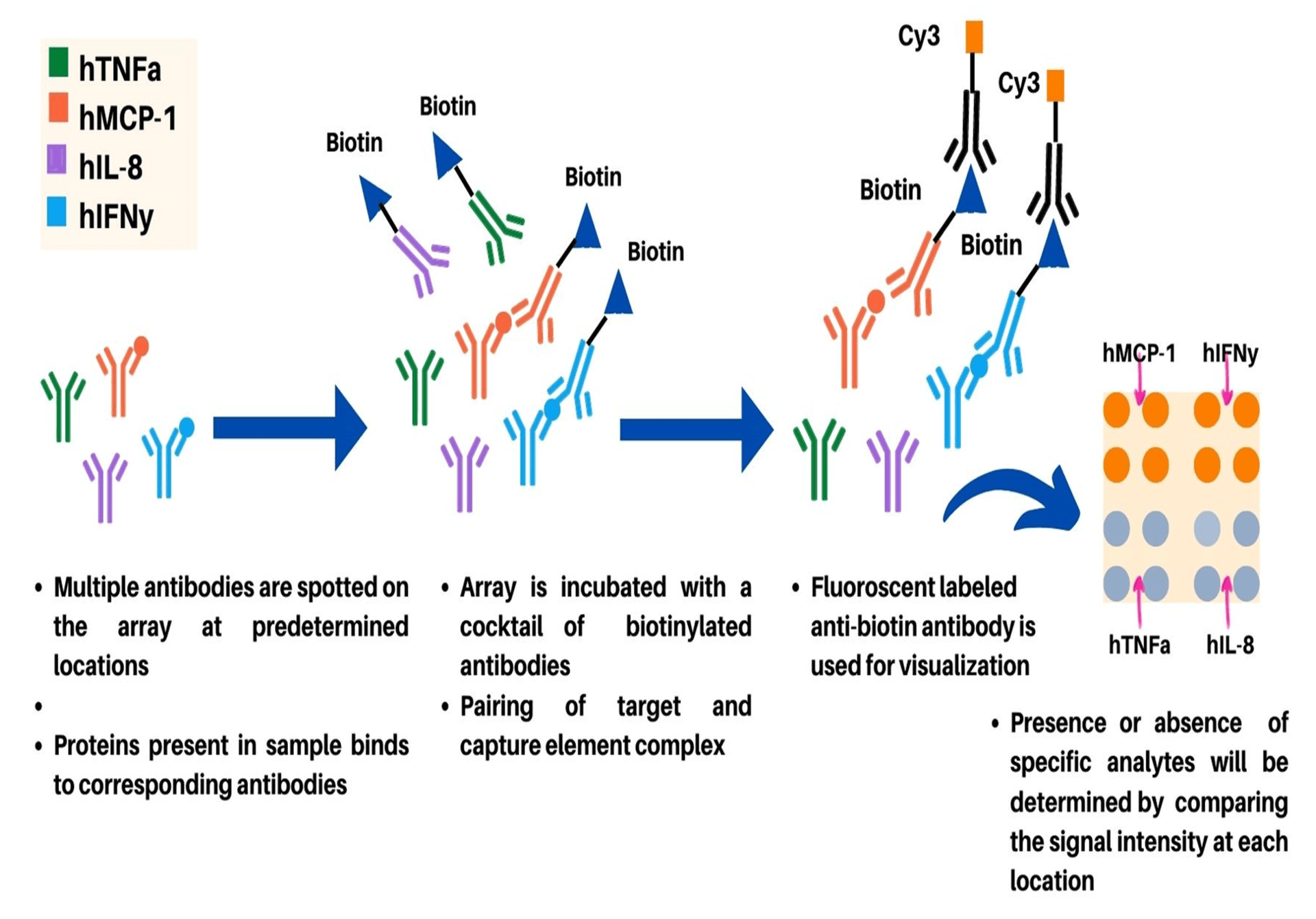
In FLISA-based antibody microarray detection format, unlabeled protein targets are captured on immobilized antibodies spotted on the solid support or chip (Figure 3). After capturing of target proteins, the array is incubated with a cocktail of biotin-labeled antibodies (primary antibody) specific to one of the spotted capture antibodies. Furthermore, incubation of array with fluorophore labeled anti-biotin secondary antibody which acts as a suitable detection antibody for visualization is carried out. The FLISA detection format provides increased sensitivity compared to the direct labeling method owing to significantly reduced background signal production in this microarray format. The sensitivity of FLISA antibody microarray detection format can be further enhanced using rolling-circle amplification or tyramide signal amplification5. However, due to difficulty in optimization of assays for measurement using various targets, FLISA detection is suitable for measurement of a limited number of targets at concentration below the detection limit of direct labeling method. As different parameters such as the affinity and specificity of the antibodies used, utilized detection conditions, and protein background in the sample are crucial for the detection limits of the assay; the FLISA method can provide detection limits in the low pg/ml range.
References
- Dubey, P.P., Kumar, D. (2013). Microarray Technology: Basic Concept, Protocols, and Applications. In: Arora, D., Das, S., Sukumar, M. (eds) Analyzing Microbes. Springer Protocols Handbooks. Springer, Berlin, Heidelberg. https://doi.org/10.1007/978-3-642-34410-7_17.
- Hoheisel JD. Microarray technology: beyond transcript profiling and genotype analysis. Nat Rev Genet. 2006 Mar;7(3):200-10. doi: 10.1038/nrg1809.
- Fung, Eric ed. Protein arrays: methods and protocols. Fremont, CA: Humana Press Inc.; 2004.
- Robinson WH, DiGennaro C, Hueber W, Haab BB, Kamachi M, Dean EJ, et al. Autoantigen microarrays for multiplex characterization of autoantibody responses. Nat Med (2002) 8:295–301. doi:10.1038/nm0302-295.
- Zhou, Heping, et al. Genome Biology, Vol. 5, issue 4, R28; 2004.
kbDNA, INC.
125 Cambridgepark Dr.
Cambridge, MA 02140
Company
Contact Us
Phone:
+1 (781) 206-2235
Fax:
+1 (781) 206-2258
Email:
info@kbDNA.com
Useful Links
The kbDNA Inc. Quality Management Network is certified as conforming to ISO 9001:2015 standard. Request Certificate